Targeted substrate degradation by Kelch controls the actin cytoskeleton during ring canal expansion
- PMID: 30559276
- PMCID: PMC6340150
- DOI: 10.1242/dev.169219
Targeted substrate degradation by Kelch controls the actin cytoskeleton during ring canal expansion
Abstract
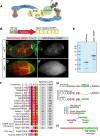
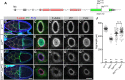
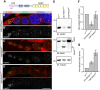
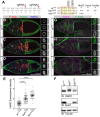
During Drosophila oogenesis, specialized actin-based structures called ring canals form and expand to accommodate growth of the oocyte. Previous work demonstrated that Kelch and Cullin 3 function together in a Cullin 3-RING ubiquitin ligase complex (CRL3Kelch) to organize the ring canal cytoskeleton, presumably by targeting a substrate for proteolysis. Here, we use tandem affinity purification followed by mass spectrometry to identify HtsRC as the CRL3Kelch ring canal substrate. CRISPR-mediated mutagenesis of HtsRC revealed its requirement in the recruitment of the ring canal F-actin cytoskeleton. We present genetic evidence consistent with HtsRC being the CRL3Kelch substrate, as well as biochemical evidence indicating that HtsRC is ubiquitylated and degraded by the proteasome. Finally, we identify a short sequence motif in HtsRC that is necessary for Kelch binding. These findings uncover an unusual mechanism during development wherein a specialized cytoskeletal structure is regulated and remodeled by the ubiquitin-proteasome system.
Keywords: Actin cytoskeleton; Cullin 3; Drosophila oogenesis; Kelch; Ring canal; Ubiquitin-proteasome system.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures



Similar articles
-
HtsRC-Mediated Accumulation of F-Actin Regulates Ring Canal Size During Drosophila melanogaster Oogenesis.Genetics. 2020 Nov;216(3):717-734. doi: 10.1534/genetics.120.303629. Epub 2020 Sep 3. Genetics. 2020. PMID: 32883702 Free PMC article.
-
Actin Cytoskeletal Organization in Drosophila Germline Ring Canals Depends on Kelch Function in a Cullin-RING E3 Ligase.Genetics. 2015 Nov;201(3):1117-31. doi: 10.1534/genetics.115.181289. Epub 2015 Sep 16. Genetics. 2015. PMID: 26384358 Free PMC article.
-
Drosophila Kelch functions with Cullin-3 to organize the ring canal actin cytoskeleton.J Cell Biol. 2010 Jan 11;188(1):29-37. doi: 10.1083/jcb.200909017. J Cell Biol. 2010. PMID: 20065088 Free PMC article.
-
The BACK domain in BTB-kelch proteins.Trends Biochem Sci. 2004 Dec;29(12):634-7. doi: 10.1016/j.tibs.2004.10.003. Trends Biochem Sci. 2004. PMID: 15544948 Review.
-
Coordinated Actions Between p97 and Cullin-RING Ubiquitin Ligases for Protein Degradation.Adv Exp Med Biol. 2020;1217:61-78. doi: 10.1007/978-981-15-1025-0_5. Adv Exp Med Biol. 2020. PMID: 31898222 Review.
Cited by
-
Complementary Volume Electron Microscopy-based approaches reveal ultrastructural changes in germline intercellular bridges of D. melanogaster.bioRxiv [Preprint]. 2025 Feb 23:2025.02.18.638836. doi: 10.1101/2025.02.18.638836. bioRxiv. 2025. PMID: 40027623 Free PMC article. Preprint.
-
HtsRC-Mediated Accumulation of F-Actin Regulates Ring Canal Size During Drosophila melanogaster Oogenesis.Genetics. 2020 Nov;216(3):717-734. doi: 10.1534/genetics.120.303629. Epub 2020 Sep 3. Genetics. 2020. PMID: 32883702 Free PMC article.
-
Tissue-specific dynamic codon redefinition in Drosophila.Proc Natl Acad Sci U S A. 2021 Feb 2;118(5):e2012793118. doi: 10.1073/pnas.2012793118. Proc Natl Acad Sci U S A. 2021. PMID: 33500350 Free PMC article.
-
Ring canals in the larval adipose of Drosophila buffer stress response.bioRxiv [Preprint]. 2025 Jun 11:2025.06.11.658881. doi: 10.1101/2025.06.11.658881. bioRxiv. 2025. PMID: 40661459 Free PMC article. Preprint.
-
Precise levels of the Drosophila adaptor protein Dreadlocks maintain the size and stability of germline ring canals.J Cell Sci. 2021 Apr 15;134(8):jcs254730. doi: 10.1242/jcs.254730. Epub 2021 Apr 27. J Cell Sci. 2021. PMID: 33912915 Free PMC article.
References
-
- Bomont P., Cavalier L., Blondeau F., Ben Hamida C., Belal S., Tazir M., Demir E., Topaloglu H., Korinthenberg R., Tüysüz B. et al. (2000). The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat. Genet. 26, 370-374. 10.1038/81701 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

