Diagnosing rare diseases after the exome
- PMID: 30559314
- PMCID: PMC6318767
- DOI: 10.1101/mcs.a003392
Diagnosing rare diseases after the exome
Abstract
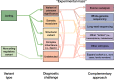
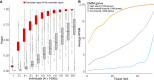
High-throughput sequencing has ushered in a diversity of approaches for identifying genetic variants and understanding genome structure and function. When applied to individuals with rare genetic diseases, these approaches have greatly accelerated gene discovery and patient diagnosis. Over the past decade, exome sequencing has emerged as a comprehensive and cost-effective approach to identify pathogenic variants in the protein-coding regions of the genome. However, for individuals in whom exome-sequencing fails to identify a pathogenic variant, we discuss recent advances that are helping to reduce the diagnostic gap.
© 2018 Frésard and Montgomery; Published by Cold Spring Harbor Laboratory Press.
Figures
Similar articles
-
Implementation of Exome Sequencing to Identify Rare Genetic Diseases.Methods Mol Biol. 2024;2719:79-98. doi: 10.1007/978-1-0716-3461-5_5. Methods Mol Biol. 2024. PMID: 37803113 Review.
-
A Diagnosis for All Rare Genetic Diseases: The Horizon and the Next Frontiers.Cell. 2019 Mar 21;177(1):32-37. doi: 10.1016/j.cell.2019.02.040. Cell. 2019. PMID: 30901545
-
Challenges in the diagnosis and discovery of rare genetic disorders using contemporary sequencing technologies.Brief Funct Genomics. 2020 Jul 29;19(4):243-258. doi: 10.1093/bfgp/elaa009. Brief Funct Genomics. 2020. PMID: 32393978 Review.
-
Massively Parallel Sequencing for Rare Genetic Disorders: Potential and Pitfalls.Front Endocrinol (Lausanne). 2021 Feb 19;11:628946. doi: 10.3389/fendo.2020.628946. eCollection 2020. Front Endocrinol (Lausanne). 2021. PMID: 33679611 Free PMC article. Review.
-
Targeted Next-Generation Sequencing in Rare Diseases.Methods Mol Biol. 2025;2866:45-57. doi: 10.1007/978-1-0716-4192-7_3. Methods Mol Biol. 2025. PMID: 39546196
Cited by
-
Targeted long-read sequencing identifies missing disease-causing variation.Am J Hum Genet. 2021 Aug 5;108(8):1436-1449. doi: 10.1016/j.ajhg.2021.06.006. Epub 2021 Jul 2. Am J Hum Genet. 2021. PMID: 34216551 Free PMC article.
-
Broadening the Genetic Spectrum of Painful Small-Fiber Neuropathy through Whole-Exome Study in Early-Onset Cases.Int J Mol Sci. 2024 Jun 30;25(13):7248. doi: 10.3390/ijms25137248. Int J Mol Sci. 2024. PMID: 39000354 Free PMC article.
-
Diagnostic Challenges of Neuromuscular Disorders after Whole Exome Sequencing.J Neuromuscul Dis. 2023;10(4):667-684. doi: 10.3233/JND-230013. J Neuromuscul Dis. 2023. PMID: 37066920 Free PMC article.
-
How to choose a test for prenatal genetic diagnosis: a practical overview.Am J Obstet Gynecol. 2023 Feb;228(2):178-186. doi: 10.1016/j.ajog.2022.08.039. Epub 2022 Aug 24. Am J Obstet Gynecol. 2023. PMID: 36029833 Free PMC article.
-
Network analysis reveals rare disease signatures across multiple levels of biological organization.Nat Commun. 2021 Nov 9;12(1):6306. doi: 10.1038/s41467-021-26674-1. Nat Commun. 2021. PMID: 34753928 Free PMC article.
References
-
- Birgmeier J, Haeussler M, Deisseroth CA, Jagadeesh KA, Ratner AJ, Guturu H, Wenger AM, Stenson PD, Cooper DN, Re C, et al. 2017. AMELIE accelerates Mendelian patient diagnosis directly from the primary literature. bioRxiv 10.1101/171322. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
