Myopathy associated BAG3 mutations lead to protein aggregation by stalling Hsp70 networks
- PMID: 30559338
- PMCID: PMC6297355
- DOI: 10.1038/s41467-018-07718-5
Myopathy associated BAG3 mutations lead to protein aggregation by stalling Hsp70 networks
Abstract
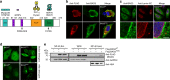
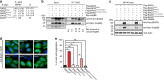
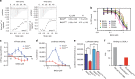
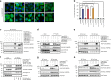
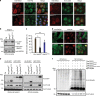
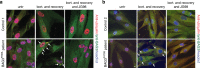
BAG3 is a multi-domain hub that connects two classes of chaperones, small heat shock proteins (sHSPs) via two isoleucine-proline-valine (IPV) motifs and Hsp70 via a BAG domain. Mutations in either the IPV or BAG domain of BAG3 cause a dominant form of myopathy, characterized by protein aggregation in both skeletal and cardiac muscle tissues. Surprisingly, for both disease mutants, impaired chaperone binding is not sufficient to explain disease phenotypes. Recombinant mutants are correctly folded, show unaffected Hsp70 binding but are impaired in stimulating Hsp70-dependent client processing. As a consequence, the mutant BAG3 proteins become the node for a dominant gain of function causing aggregation of itself, Hsp70, Hsp70 clients and tiered interactors within the BAG3 interactome. Importantly, genetic and pharmaceutical interference with Hsp70 binding completely reverses stress-induced protein aggregation for both BAG3 mutations. Thus, the gain of function effects of BAG3 mutants act as Achilles heel of the HSP70 machinery.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Kampinga, H. H. & Bergink, S. Heat shock proteins as potential targets for protective strategies in neurodegeneration. Lancet Neurol.15, 748–759 (2016) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

