A rapid and robust method for single cell chromatin accessibility profiling
- PMID: 30559361
- PMCID: PMC6297232
- DOI: 10.1038/s41467-018-07771-0
A rapid and robust method for single cell chromatin accessibility profiling
Abstract
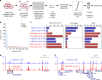
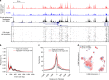
The assay for transposase-accessible chromatin using sequencing (ATAC-seq) is widely used to identify regulatory regions throughout the genome. However, very few studies have been performed at the single cell level (scATAC-seq) due to technical challenges. Here we developed a simple and robust plate-based scATAC-seq method, combining upfront bulk Tn5 tagging with single-nuclei sorting. We demonstrate that our method works robustly across various systems, including fresh and cryopreserved cells from primary tissues. By profiling over 3000 splenocytes, we identify distinct immune cell types and reveal cell type-specific regulatory regions and related transcription factors.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

