Complete genome analysis of a Siphoviridae phage TSK1 showing biofilm removal potential against Klebsiella pneumoniae
- PMID: 30559386
- PMCID: PMC6297243
- DOI: 10.1038/s41598-018-36229-y
Complete genome analysis of a Siphoviridae phage TSK1 showing biofilm removal potential against Klebsiella pneumoniae
Abstract
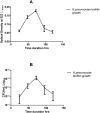
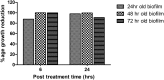
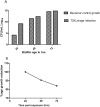
Multidrug-resistant Klebsiella pneumoniae is a nosocomial pathogen, produces septicemia, pneumonia and UTI. Excessive use of antibiotics contributes towards emergence of multidrug-resistance. Bacteriophage-therapy is a potential substitute of antibiotics with many advantages. In this investigation, microbiological and genome characterization of TSK1 bacteriophage and its biofilm elimination capability are presented. TSK1 showed narrow host range and highest stability at pH 7 and 37 °C. TSK1 reduced the growth of K. pneumoniae during the initial 14 hours of infection. Post-treatment with TSK1 against different age K. pneumoniae biofilms reduced 85-100% biomass. Pre-treatment of TSK1 bacteriophage against the biofilm of Klebsiella pneumoniae reduced > 99% biomass in initial 24 hr of incubation. The genome of TSK1 phage comprised 49,836 base pairs with GC composition of 50.44%. Total seventy-five open reading frames (ORFs) were predicted, 25 showed homology with known functional proteins, while 50 were called hypothetical, as no homologs with proved function exists in the genome databases. Blast and phylogenetic analysis put it in the Kp36 virus genus of family Siphoviridae. Proposed packaging strategy of TSK1 bacteriophage genome is headful packaging using the pac sites. The potential of TSK1 bacteriophage could be used to reduce the bacterial load and biofilm in clinical and non-clinical settings.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Saleem AF, Qamar FN, Shahzad H, Qadir M, Zaidi AK. Trends in antibiotic susceptibility and incidence of late-onset Klebsiella pneumoniae neonatal sepsis over a six-year period in a neonatal intensive care unit in Karachi, Pakistan. Int J Infect Dis. 2013;17:e961–e965. doi: 10.1016/j.ijid.2013.04.007. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 4501/Higher Education Commission, Pakistan (HEC)/International
- 4501/Higher Education Commission, Pakistan (HEC)/International
- 4501/Higher Education Commission, Pakistan (HEC)/International
- 4501/Higher Education Commission, Pakistan (HEC)/International
- 4501/Alabama Commission on Higher Education/International
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

