Imaging cellular ultrastructures using expansion microscopy (U-ExM)
- PMID: 30559430
- PMCID: PMC6314451
- DOI: 10.1038/s41592-018-0238-1
Imaging cellular ultrastructures using expansion microscopy (U-ExM)
Abstract
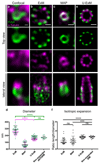
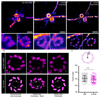
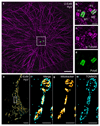
Determining the structure and composition of macromolecular assemblies is a major challenge in biology. Here we describe ultrastructure expansion microscopy (U-ExM), an extension of expansion microscopy that allows the visualization of preserved ultrastructures by optical microscopy. This method allows for near-native expansion of diverse structures in vitro and in cells; when combined with super-resolution microscopy, it unveiled details of ultrastructural organization, such as centriolar chirality, that could otherwise be observed only by electron microscopy.
Conflict of interest statement
Authors declare no competing interests.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

