HCV Defective Genomes Promote Persistent Infection by Modulating the Viral Life Cycle
- PMID: 30559733
- PMCID: PMC6287115
- DOI: 10.3389/fmicb.2018.02942
HCV Defective Genomes Promote Persistent Infection by Modulating the Viral Life Cycle
Abstract
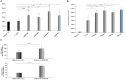
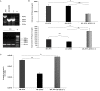
Defective interfering (DI) RNAs have been detected in several human viruses. HCV in-frame deletions mutants (IFDMs), missing mainly the envelope proteins, have been found in patient sera and liver tissues. IFDMs replicate independently and can be trans-packaged into infectious virions in the presence of full length viral genome. So far, their biological role is unclear. In this study, we have isolated and cloned IFDMs from sera samples and liver tissues of patients infected with HCV genotypes 1b, 2a, and 3a. IFDMs were present in up to 26% of samples tested. Using the in vitro HCV cell culture system, co-expression of the wild type (wt) HCV replicon with HCV IFDMs RNA resulted in increased HCV replication. Additionally, co-transfection of the HCV full length genome RNA and a defective mutant missing the envelope region led to increased viral release, collectively suggesting an important biological role for IFDMs in the virus life cycle. Recently, exosomes, masters of intercellular communication, have been implicated in the transport of HCV viral genomes. We report for the first time that exosomal RNA isolated from HCV sera samples contains HCV defective genomes. We also demonstrate that inhibition of exosomal biogenesis and release influences HCV viral replication. Overall, we provide evidence that the presence of HCV IFDMs affects both viral replication and release. IFDMs exploit exosomes as means of transport, a way to evade the immune system, to spread more efficiently and possibly maintain persistent infection.
Keywords: defective genomes; exosomes; hepatitis C; viral persistence; viral replication.
Figures


References
-
- Cheroni C., Donnici L., Aghemo A., Balistreri F., Bianco A., Zanoni V., et al. (2015). Hepatitis C Virus deletion mutants are found in individuals chronically infected with genotype 1 hepatitis C virus in association with age, high viral load and liver inflammatory activity. PLoS One 10:e0138546. 10.1371/journal.pone.0138546 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

