Zoonotic Source Attribution of Salmonella enterica Serotype Typhimurium Using Genomic Surveillance Data, United States
- PMID: 30561314
- PMCID: PMC6302586
- DOI: 10.3201/eid2501.180835
Zoonotic Source Attribution of Salmonella enterica Serotype Typhimurium Using Genomic Surveillance Data, United States
Abstract
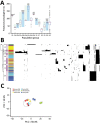
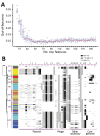
Increasingly, routine surveillance and monitoring of foodborne pathogens using whole-genome sequencing is creating opportunities to study foodborne illness epidemiology beyond routine outbreak investigations and case-control studies. Using a global phylogeny of Salmonella enterica serotype Typhimurium, we found that major livestock sources of the pathogen in the United States can be predicted through whole-genome sequencing data. Relatively steady rates of sequence divergence in livestock lineages enabled the inference of their recent origins. Elevated accumulation of lineage-specific pseudogenes after divergence from generalist populations and possible metabolic acclimation in a representative swine isolate indicates possible emergence of host adaptation. We developed and retrospectively applied a machine learning Random Forest classifier for genomic source prediction of Salmonella Typhimurium that correctly attributed 7 of 8 major zoonotic outbreaks in the United States during 1998-2013. We further identified 50 key genetic features that were sufficient for robust livestock source prediction.
Keywords: Salmonella; Salmonella enterica serotype Typhimurium; United States; bacteria; machine learning; population structure; source attribution; whole-genome sequencing; zoonoses.
Figures


References
-
- Hoffmann S, Maculloch B, Batz M. Economic burden of major foodborne illnesses acquired in the United States [cited 2018 Oct 8]. https://www.ers.usda.gov/webdocs/publications/43984/52807_eib140.pdf
-
- Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, et al. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis. 2011;8:887–900. 10.1089/fpd.2010.0787 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

