Human Immunodeficiency Virus Type 1 RNA Detected in the Central Nervous System (CNS) After Years of Suppressive Antiretroviral Therapy Can Originate from a Replicating CNS Reservoir or Clonally Expanded Cells
- PMID: 30561541
- PMCID: PMC6938202
- DOI: 10.1093/cid/ciy1066
Human Immunodeficiency Virus Type 1 RNA Detected in the Central Nervous System (CNS) After Years of Suppressive Antiretroviral Therapy Can Originate from a Replicating CNS Reservoir or Clonally Expanded Cells
Abstract
Background: Human immunodeficiency virus type 1 (HIV-1) populations are detected in cerebrospinal fluid (CSF) of some people on suppressive antiretroviral therapy (ART). Detailed analysis of these populations may reveal whether they are produced by central nervous system (CNS) reservoirs.
Methods: We performed a study of 101 asymptomatic participants on stable ART. HIV-1 RNA concentrations were cross-sectionally measured in CSF and plasma. In participants with CSF HIV-1 RNA concentrations sufficient for analysis, viral populations were genetically and phenotypically characterized over multiple time points.
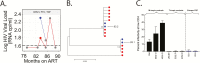
Results: For 6% of participants (6 of 101), the concentration of HIV-1 RNA in their CSF was ≥0.5 log copies/mL above that of plasma (ie, CSF escape). We generated viral envelope sequences from CSF of 3 participants. One had a persistent CSF escape population that was macrophage-tropic, partially drug resistant, genetically diverse, and closely related to a minor macrophage-tropic lineage present in the blood prior to viral suppression and enriched for after ART. Two participants (1 suppressed and 1 not) had transient CSF escape populations that were R5 T cell-tropic with little genetic diversity.
Conclusions: Extensive analysis of viral populations in 1 participant revealed that CSF escape was from a persistently replicating population, likely in macrophages/microglia, present in the CNS over 3 years of ART. CSF escape in 2 other participants was likely produced by trafficking and transient expansion of infected T cells in the CNS. Our results show that CNS reservoirs can persist during ART and that CSF escape is not exclusively produced by replicating CNS reservoirs.
Keywords: CNS; CSF escape; HIV reservoirs; drug resistance; persistence.
© The Author(s) 2018. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures



References
-
- Doyle T, Smith C, Vitiello P, et al. . Plasma HIV-1 RNA detection below 50 copies/ml and risk of virologic rebound in patients receiving highly active antiretroviral therapy. Clin Infect Dis 2012; 54:724–32. - PubMed
-
- Hermankova M, Ray SC, Ruff C, et al. . HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 2001; 286:196–207. - PubMed
-
- Kieffer TL, Finucane MM, Nettles RE, et al. . Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis 2004; 189:1452–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

