A Divergent Hepatitis D-Like Agent in Birds
- PMID: 30562970
- PMCID: PMC6315422
- DOI: 10.3390/v10120720
A Divergent Hepatitis D-Like Agent in Birds
Abstract
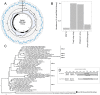
Hepatitis delta virus (HDV) is currently only found in humans and is a satellite virus that depends on hepatitis B virus (HBV) envelope proteins for assembly, release, and entry. Using meta-transcriptomics, we identified the genome of a novel HDV-like agent in ducks. Sequence analysis revealed secondary structures that were shared with HDV, including self-complementarity and ribozyme features. The predicted viral protein shares 32% amino acid similarity to the small delta antigen of HDV and comprises a divergent phylogenetic lineage. The discovery of an avian HDV-like agent has important implications for the understanding of the origins of HDV and sub-viral agents.
Keywords: co-evolution; dabbling duck; hepatitis D virus; phylogeny.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Taylor J.M., Purcell R.H., Farci P. Hepatitis D (D) virus. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 2222–2241.
-
- WHO Hepatitis D Fact Sheet. [(accessed on 24 February 2018)];2017 Available online: http://www.who.int/mediacentre/factsheets/hepatitis-d/en/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

