The Dictyostelium discoideum homologue of Twinkle, Twm1, is a mitochondrial DNA helicase, an active primase and promotes mitochondrial DNA replication
- PMID: 30563453
- PMCID: PMC6299598
- DOI: 10.1186/s12867-018-0114-7
The Dictyostelium discoideum homologue of Twinkle, Twm1, is a mitochondrial DNA helicase, an active primase and promotes mitochondrial DNA replication
Abstract
Background: DNA replication requires contributions from various proteins, such as DNA helicases; in mitochondria Twinkle is important for maintaining and replicating mitochondrial DNA. Twinkle helicases are predicted to also possess primase activity, as has been shown in plants; however this activity appears to have been lost in metazoans. Given this, the study of Twinkle in other organisms is required to better understand the evolution of this family and the roles it performs within mitochondria.
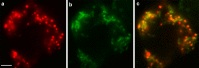
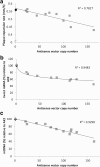
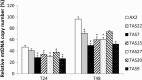
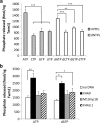
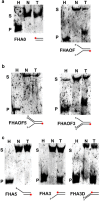
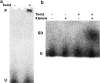
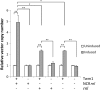
Results: Here we describe the characterization of a Twinkle homologue, Twm1, in the amoeba Dictyostelium discoideum, a model organism for mitochondrial genetics and disease. We show that Twm1 is important for mitochondrial function as it maintains mitochondrial DNA copy number in vivo. Twm1 is a helicase which unwinds DNA resembling open forks, although it can act upon substrates with a single 3' overhang, albeit less efficiently. Furthermore, unlike human Twinkle, Twm1 has primase activity in vitro. Finally, using a novel in bacterio approach, we demonstrated that Twm1 promotes DNA replication.
Conclusions: We conclude that Twm1 is a replicative mitochondrial DNA helicase which is capable of priming DNA for replication. Our results also suggest that non-metazoan Twinkle could function in the initiation of mitochondrial DNA replication. While further work is required, this study has illuminated several alternative processes of mitochondrial DNA maintenance which might also be performed by the Twinkle family of helicases.
Keywords: DNA helicase; DNA primase; Dictyostelium discoideum; Mitochondrial DNA replication; Twinkle.
Figures

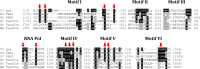
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

