Genome Mining of Plant NPFs Reveals Varying Conservation of Signature Motifs Associated With the Mechanism of Transport
- PMID: 30564251
- PMCID: PMC6288477
- DOI: 10.3389/fpls.2018.01668
Genome Mining of Plant NPFs Reveals Varying Conservation of Signature Motifs Associated With the Mechanism of Transport
Abstract
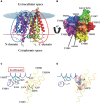
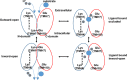
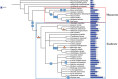
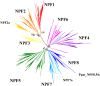
Nitrogen is essential for all living species and may be taken up from the environment in different forms like nitrate or peptides. In plants, members of a transporter family named NPFs transport nitrate and peptides across biological membranes. NPFs are phylogenetically related to a family of peptide transporters (PTRs) or proton-coupled oligopeptide transporters (POTs) that are evolutionarily conserved in all organisms except in Archaea. POTs are present in low numbers in bacteria, algae and animals. NPFs have expanded in plants and evolved to transport a wide range of substrates including phytohormones and glucosinolates. Functional studies have shown that most NPFs, like POTs, operate as symporters with simultaneous inwardly directed movement of protons. Here we focus on four structural features of NPFs/POTs/PTRs that have been shown by structural and functional studies to be essential to proton-coupled symport transport. The first two features are implicated in proton binding and transport: a conserved motif named ExxER/K, located in the first transmembrane helix (TMH1) and a D/E residue in TMH7 that has been observed in some bacterial and algal transporters. The third and fourth features are two inter-helical salt bridges between residues on TMH1 and TMH7 or TMH4 and TMH10. To understand if the mechanism of transport is conserved in NPFs with the expansion to novel substrates, we collected NPFs sequences from 42 plant genomes. Sequence alignment revealed that the ExxER/K motif is not strictly conserved and its conservation level is different in the NPF subfamilies. The proton binding site on TMH7 is missing in all NPFs with the exception of two NPFs from moss. The two moss NPFs also have a positively charged amino acid on TMH1 that can form the salt bridge with the TMH7 negative residue. None of the other NPFs we examined harbor residues that can form the TMH1-TMH7 salt bridge. In contrast, the amino acids required to form the TMH4-TMH10 salt bridge are highly conserved in NPFs, with some exceptions. These results support the need for further biochemical and structural studies of individual NPFs for a better understanding of the transport mechanism in this family of transporters.
Keywords: genome; gibberellin); glucosinolates (GSL); nitrate; nitrate peptide family (NPF) transporters; nitrogen; phytohormones (auxin; proton-dependent oligopeptide transporter.
Figures


References
LinkOut - more resources
Full Text Sources

