Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients
- PMID: 30564623
- PMCID: PMC6292381
- DOI: 10.1002/acn3.649
Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients
Abstract
Objective: Limb-girdle muscular dystrophies (LGMDs), one of the most heterogeneous neuromuscular disorders (NMDs), involves predominantly proximal-muscle weakness with >30 genes associated with different subtypes. The clinical-genetic overlap among subtypes and with other NMDs complicate disease-subtype identification lengthening diagnostic process, increases overall costs hindering treatment/clinical-trial recruitment. Currently seven LGMD clinical trials are active but still no gene-therapy-related treatment is available. Till-date no nation-wide large-scale LGMD sequencing program was performed. Our objectives were to understand LGMD genetic basis, different subtypes' relative prevalence across US and investigate underlying disease mechanisms.
Methods: A total of 4656 patients with clinically suspected-LGMD across US were recruited to conduct next-generation sequencing (NGS)-based gene-panel testing during June-2015 to June-2017 in CLIA-CAP-certified Emory-Genetics-Laboratory. Thirty-five LGMD-subtypes-associated or LGMD-like other NMD-associated genes were investigated. Main outcomes were diagnostic yield, gene-variant spectrum, and LGMD subtypes' prevalence in a large US LGMD-suspected population.
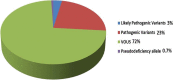
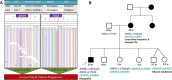
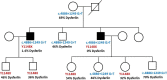
Results: Molecular diagnosis was established in 27% (1259 cases; 95% CI, 26-29%) of the patients with major contributing genes to LGMD phenotypes being: CAPN3(17%), DYSF(16%), FKRP(9%) and ANO5(7%). We observed an increased prevalence of genetically confirmed late-onset Pompe disease, DNAJB6-associated LGMD subtype1E and CAPN3-associated autosomal-dominant LGMDs. Interestingly, we identified a high prevalence of patients with pathogenic variants in more than one LGMD gene suggesting possible synergistic heterozygosity/digenic/multigenic contribution to disease presentation/progression that needs consideration as a part of diagnostic modality.
Interpretation: Overall, this study has improved our understanding of the relative prevalence of different LGMD subtypes, their respective genetic etiology, and the changing paradigm of their inheritance modes and novel mechanisms that will allow for improved timely treatment, management, and enrolment of molecularly diagnosed individuals in clinical trials.
Figures


Comment in
-
Reply: Autosomal dominant segregation of CAPN3 c.598_612del15 associated with a mild form of calpainopathy.Ann Clin Transl Neurol. 2020 Dec;7(12):2541. doi: 10.1002/acn3.51192. Epub 2020 Oct 15. Ann Clin Transl Neurol. 2020. PMID: 33058423 Free PMC article. No abstract available.
-
Autosomal dominant segregation of CAPN3 c.598_612del15 associated with a mild form of calpainopathy.Ann Clin Transl Neurol. 2020 Dec;7(12):2538-2540. doi: 10.1002/acn3.51193. Epub 2020 Oct 27. Ann Clin Transl Neurol. 2020. PMID: 33107701 Free PMC article. No abstract available.
References
-
- Fanin M, Angelini C. Progress and challenges in diagnosis of dysferlinopathy. Muscle Nerve 2016;54:821–835. - PubMed
-
- Bushby KM. The limb‐girdle muscular dystrophies‐multiple genes, multiple mechanisms. Hum Mol Genet 1999;8:1875–1882. - PubMed
-
- Guglieri M, Magri F, D'Angelo MG, et al. Clinical, molecular, and protein correlations in a large sample of genetically diagnosed Italian limb girdle muscular dystrophy patients. Hum Mutat 2008;29:258–266. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

