Selective Export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation
- PMID: 30566862
- PMCID: PMC6392044
- DOI: 10.1016/j.celrep.2018.11.073
Selective Export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation
Abstract
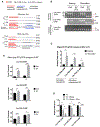
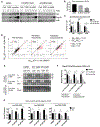
The discovery of microRNA (miRNA) sorting into extracellular vesicles (EVs) revealed a novel mode of intercellular communication and uncovered a link between cellular endomembrane compartments and small RNAs in EV-secreting cells. Using a two-step ultracentrifugation procedure to isolate EVs released by T cells, we found that 45% of tRNA fragments (tRFs), but fewer than 1% of miRNAs, were significantly enriched in EVs compared with the corresponding cellular RNA. T cell activation induced the EV-mediated release of a specific set of tRFs derived from the 5' end and 3'-internal region of tRNAs without variable loops. Inhibition of EV biogenesis pathways specifically led to the accumulation of these activation-induced EV-enriched tRFs within multivesicular bodies (MVBs). Introducing antisense oligonucleotides to inhibit these tRFs enhanced T cell activation. Taken together, these results demonstrate that T cells selectively release tRFs into EVs via MVBs and suggest that this process may remove tRFs that repress immune activation.
Keywords: T lymphocyte; exosome; extracellular vesicle; tRNA fragment; tsRNA.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures





References
-
- Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Pérez Lanzón M, Zini N, Naaijkens B, Perut F, Niessen HW, Baldini N, and Pegtel DM (2015). Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther 6, 127. - PMC - PubMed
-
- Barcena C, Stefanovic M, Tutusaus A, Martinez-Nieto GA, Martinez L, Garcia-Ruiz C, de Mingo A, Caballeria J, Fernandez-Checa JC, Mari M, and Morales A (2015). Angiogenin secretion from hepatoma cells activates hepatic stellate cells to amplify a self-sustained cycle promoting liver cancer. Sci. Rep 5, 7916. - PMC - PubMed
-
- Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, and Waalkes TP (1977). High turnover rate of transfer RNA in tumor tissue. Cancer Res 37, 3362–3366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

