Single-Cell Analysis Identifies LY6D as a Marker Linking Castration-Resistant Prostate Luminal Cells to Prostate Progenitors and Cancer
- PMID: 30566873
- PMCID: PMC6315111
- DOI: 10.1016/j.celrep.2018.11.069
Single-Cell Analysis Identifies LY6D as a Marker Linking Castration-Resistant Prostate Luminal Cells to Prostate Progenitors and Cancer
Abstract
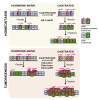
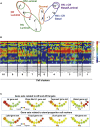
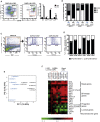
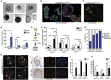
The exact identity of castrate-resistant (CR) cells and their relation to CR prostate cancer (CRPC) is unresolved. We use single-cell gene profiling to analyze the molecular heterogeneity in basal and luminal compartments. Within the luminal compartment, we identify a subset of cells intrinsically resistant to castration with a bi-lineage gene expression pattern. We discover LY6D as a marker of CR prostate progenitors with multipotent differentiation and enriched organoid-forming capacity. Lineage tracing further reveals that LY6D+ CR luminal cells can produce LY6D- luminal cells. In contrast, in luminal cells lacking PTEN, LY6D+ cells predominantly give rise to LY6D+ tumor cells, contributing to high-grade PIN lesions. Gene expression analyses in patients' biopsies indicate that LY6D expression correlates with early disease progression, including progression to CRPC. Our studies thus identify a subpopulation of luminal progenitors characterized by LY6D expression and intrinsic castration resistance. LY6D may serve as a prognostic maker for advanced prostate cancer.
Keywords: LY6D; castration-resistant prostate cancer; intrinsic resistance; luminal progenitor; tumor-initiating cells.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Brakenhoff R.H., Gerretsen M., Knippels E.M., van Dijk M., van Essen H., Weghuis D.O., Sinke R.J., Snow G.B., van Dongen G.A. The human E48 antigen, highly homologous to the murine Ly-6 antigen ThB, is a GPI-anchored molecule apparently involved in keratinocyte cell-cell adhesion. J. Cell Biol. 1995;129:1677–1689. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

