Combining multiple functional annotation tools increases coverage of metabolic annotation
- PMID: 30567498
- PMCID: PMC6299973
- DOI: 10.1186/s12864-018-5221-9
Combining multiple functional annotation tools increases coverage of metabolic annotation
Abstract
Background: Genome-scale metabolic modeling is a cornerstone of systems biology analysis of microbial organisms and communities, yet these genome-scale modeling efforts are invariably based on incomplete functional annotations. Annotated genomes typically contain 30-50% of genes without functional annotation, severely limiting our knowledge of the "parts lists" that the organisms have at their disposal. These incomplete annotations may be sufficient to derive a model of a core set of well-studied metabolic pathways that support growth in pure culture. However, pathways important for growth on unusual metabolites exchanged in complex microbial communities are often less understood, resulting in missing functional annotations in newly sequenced genomes.
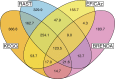
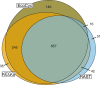
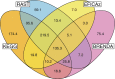
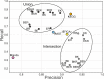
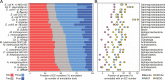
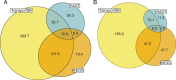
Results: Here, we present results on a comprehensive reannotation of 27 bacterial reference genomes, focusing on enzymes with EC numbers annotated by KEGG, RAST, EFICAz, and the BRENDA enzyme database, and on membrane transport annotations by TransportDB, KEGG and RAST. Our analysis shows that annotation using multiple tools can result in a drastically larger metabolic network reconstruction, adding on average 40% more EC numbers, 3-8 times more substrate-specific transporters, and 37% more metabolic genes. These results are even more pronounced for bacterial species that are phylogenetically distant from well-studied model organisms such as E. coli.
Conclusions: Metabolic annotations are often incomplete and inconsistent. Combining multiple functional annotation tools can greatly improve genome coverage and metabolic network size, especially for non-model organisms and non-core pathways.
Keywords: Enzyme prediction; Functional annotation; Genome annotation; Metabolic modeling; Transport prediction.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
High-throughput comparison, functional annotation, and metabolic modeling of plant genomes using the PlantSEED resource.Proc Natl Acad Sci U S A. 2014 Jul 1;111(26):9645-50. doi: 10.1073/pnas.1401329111. Epub 2014 Jun 9. Proc Natl Acad Sci U S A. 2014. PMID: 24927599 Free PMC article.
-
High precision multi-genome scale reannotation of enzyme function by EFICAz.BMC Genomics. 2006 Dec 13;7:315. doi: 10.1186/1471-2164-7-315. BMC Genomics. 2006. PMID: 17166279 Free PMC article.
-
Modeling central metabolism and energy biosynthesis across microbial life.BMC Genomics. 2016 Aug 8;17:568. doi: 10.1186/s12864-016-2887-8. BMC Genomics. 2016. PMID: 27502787 Free PMC article.
-
Using comparative genome analysis to identify problems in annotated microbial genomes.Microbiology (Reading). 2010 Jul;156(Pt 7):1909-1917. doi: 10.1099/mic.0.033811-0. Epub 2010 Apr 29. Microbiology (Reading). 2010. PMID: 20430813 Review.
-
Comparative Genome Annotation.Methods Mol Biol. 2018;1704:189-212. doi: 10.1007/978-1-4939-7463-4_6. Methods Mol Biol. 2018. PMID: 29277866 Review.
Cited by
-
Prokaryotic Genome Annotation.Methods Mol Biol. 2022;2349:193-214. doi: 10.1007/978-1-0716-1585-0_10. Methods Mol Biol. 2022. PMID: 34718997
-
An assessment of genome annotation coverage across the bacterial tree of life.Microb Genom. 2020 Mar;6(3):e000341. doi: 10.1099/mgen.0.000341. Microb Genom. 2020. PMID: 32124724 Free PMC article.
-
Application of the omics sciences to the study of Naegleria fowleri, Acanthamoeba spp., and Balamuthia mandrillaris: current status and future projections.Parasite. 2021;28:36. doi: 10.1051/parasite/2021033. Epub 2021 Apr 12. Parasite. 2021. PMID: 33843581 Free PMC article. Review.
-
Nutrition or nature: using elementary flux modes to disentangle the complex forces shaping prokaryote pan-genomes.BMC Ecol Evol. 2022 Aug 16;22(1):101. doi: 10.1186/s12862-022-02052-3. BMC Ecol Evol. 2022. PMID: 35974327 Free PMC article.
-
Introduction and adaptation of an emerging pathogen to olive trees in Italy.Microb Genom. 2021 Dec;7(12):000735. doi: 10.1099/mgen.0.000735. Microb Genom. 2021. PMID: 34904938 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

