Detyrosinated microtubules spatially constrain lysosomes facilitating lysosome-autophagosome fusion
- PMID: 30567713
- PMCID: PMC6363446
- DOI: 10.1083/jcb.201807124
Detyrosinated microtubules spatially constrain lysosomes facilitating lysosome-autophagosome fusion
Abstract
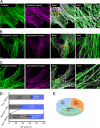
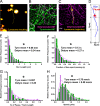
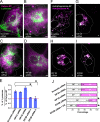
Microtubule post-translational modifications impart functional diversity to microtubules by affecting their dynamics, organization, and interaction with proteins. Using super-resolution microscopy, we show that only a small subpopulation of microtubules are detyrosinated in epithelial cells, while acetylated and tyrosinated microtubules comprise the majority of all microtubules. Surprisingly, lysosomes are enriched by approximately threefold on detyrosinated microtubules. Further, their motility on detyrosinated microtubules is impaired, showing shorter runs and more frequent and longer pauses. Lysosome enrichment is mediated through a kinesin-1-dependent mechanism, since knocking down this motor abolishes enrichment. Finally, correlative live-cell and super-resolution microscopy showed that lysosomes interact with autophagosomes on detyrosinated microtubules. Removal of detyrosinated microtubules or knockdown of kinesin-1 leads to a decrease in the percentage of autolysosomes, a fusion intermediate of autophagosomes and lysosomes. Taken together, our data reveal a new role of detyrosinated microtubules as hubs that spatially concentrate lysosomes on a small subset of microtubules and facilitate their interaction and fusion with autophagosomes to initiate autophagy.
© 2019 Mohan et al.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

