The catalytic mechanism of electron-bifurcating electron transfer flavoproteins (ETFs) involves an intermediary complex with NAD<sup/>
- PMID: 30567738
- PMCID: PMC6398123
- DOI: 10.1074/jbc.RA118.005653
The catalytic mechanism of electron-bifurcating electron transfer flavoproteins (ETFs) involves an intermediary complex with NAD<sup/>
Erratum in
-
Correction: The catalytic mechanism of electron-bifurcating electron transfer flavoproteins (ETFs) involves an intermediary complex with NAD+.J Biol Chem. 2020 Nov 6;295(45):15423. doi: 10.1074/jbc.AAC120.016322. J Biol Chem. 2020. PMID: 33158919 Free PMC article. No abstract available.
Abstract
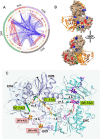
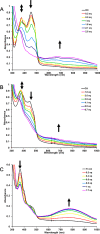
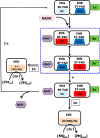
Electron bifurcation plays a key role in anaerobic energy metabolism, but it is a relatively new discovery, and only limited mechanistic information is available on the diverse enzymes that employ it. Herein, we focused on the bifurcating electron transfer flavoprotein (ETF) from the hyperthermophilic archaeon Pyrobaculum aerophilum The EtfABCX enzyme complex couples NADH oxidation to the endergonic reduction of ferredoxin and exergonic reduction of menaquinone. We developed a model for the enzyme structure by using nondenaturing MS, cross-linking, and homology modeling in which EtfA, -B, and -C each contained FAD, whereas EtfX contained two [4Fe-4S] clusters. On the basis of analyses using transient absorption, EPR, and optical titrations with NADH or inorganic reductants with and without NAD+, we propose a catalytic cycle involving formation of an intermediary NAD+-bound complex. A charge transfer signal revealed an intriguing interplay of flavin semiquinones and a protein conformational change that gated electron transfer between the low- and high-potential pathways. We found that despite a common bifurcating flavin site, the proposed EtfABCX catalytic cycle is distinct from that of the genetically unrelated bifurcating NADH-dependent ferredoxin NADP+ oxidoreductase (NfnI). The two enzymes particularly differed in the role of NAD+, the resting and bifurcating-ready states of the enzymes, how electron flow is gated, and the two two-electron cycles constituting the overall four-electron reaction. We conclude that P. aerophilum EtfABCX provides a model catalytic mechanism that builds on and extends previous studies of related bifurcating ETFs and can be applied to the large bifurcating ETF family.
Keywords: EtfABCX; anaerobic physiology; archaea; bifurcation; bioenergetics; charge transfer; electron paramagnetic resonance (EPR); electron transport; extreme thermophile; flavin; flavoprotein; radical; semiquinone.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

