Breaking the speed limit with multimode fast scanning of DNA by Endonuclease V
- PMID: 30568191
- PMCID: PMC6300609
- DOI: 10.1038/s41467-018-07797-4
Breaking the speed limit with multimode fast scanning of DNA by Endonuclease V
Erratum in
-
Publisher Correction: Breaking the speed limit with multimode fast scanning of DNA by Endonuclease V.Nat Commun. 2019 Apr 25;10(1):1991. doi: 10.1038/s41467-019-10070-x. Nat Commun. 2019. PMID: 31024006 Free PMC article.
Abstract
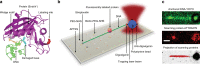
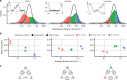
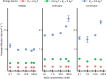
In order to preserve genomic stability, cells rely on various repair pathways for removing DNA damage. The mechanisms how enzymes scan DNA and recognize their target sites are incompletely understood. Here, by using high-localization precision microscopy along with 133 Hz high sampling rate, we have recorded EndoV and OGG1 interacting with 12-kbp elongated λ-DNA in an optical trap. EndoV switches between three distinct scanning modes, each with a clear range of activation energy barriers. These results concur with average diffusion rate and occupancy of states determined by a hidden Markov model, allowing us to infer that EndoV confinement occurs when the intercalating wedge motif is involved in rigorous probing of the DNA, while highly mobile EndoV may disengage from a strictly 1D helical diffusion mode and hop along the DNA. This makes EndoV the first example of a monomeric, single-conformation and single-binding-site protein demonstrating the ability to switch between three scanning modes.
Conflict of interest statement
The authors declare no competing interests. I.R. is currently employed by F. Hoffmann-La Roche (Roche Norge AS). The data included in this paper are based on research which has had no influence or involvement by F. Hoffmann-La Roche by any means. Any personal views of I.R. should not be understood or quoted as being made on behalf of or reflecting the position of F. Hoffmann-La Roche.
Figures

References
-
- Friedberg, E. C. et al. DNA Repair and Mutagenesis (ASM Press, Washington, 2006).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

