UK circulating strains of human parainfluenza 3: an amplicon based next generation sequencing method and phylogenetic analysis
- PMID: 30569021
- PMCID: PMC6281019
- DOI: 10.12688/wellcomeopenres.14730.2
UK circulating strains of human parainfluenza 3: an amplicon based next generation sequencing method and phylogenetic analysis
Abstract
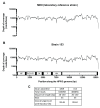
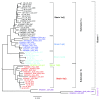
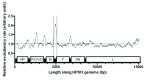
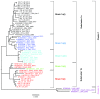
Background: Human parainfluenza viruses type 3 (HPIV3) are a prominent cause of respiratory infection with a significant impact in both pediatric and transplant patient cohorts. Currently there is a paucity of whole genome sequence data that would allow for detailed epidemiological and phylogenetic analysis of circulating strains in the UK. Although it is known that HPIV3 peaks annually in the UK, to date there are no whole genome sequences of HPIV3 UK strains available. Methods: Clinical strains were obtained from HPIV3 positive respiratory patient samples collected between 2011 and 2015. These were then amplified using an amplicon based method, sequenced on the Illumina platform and assembled using a new robust bioinformatics pipeline. Phylogenetic analysis was carried out in the context of other epidemiological studies and whole genome sequence data currently available with stringent exclusion of significantly culture-adapted strains of HPIV3. Results: In the current paper we have presented twenty full genome sequences of UK circulating strains of HPIV3 and a detailed phylogenetic analysis thereof. We have analysed the variability along the HPIV3 genome and identified a short hypervariable region in the non-coding segment between the M (matrix) and F (fusion) genes. The epidemiological classifications obtained by using this region and whole genome data were then compared and found to be identical. Conclusions: The majority of HPIV3 strains were observed at different geographical locations and with a wide temporal spread, reflecting the global distribution of HPIV3. Consistent with previous data, a particular subcluster or strain was not identified as specific to the UK, suggesting that a number of genetically diverse strains circulate at any one time. A small hypervariable region in the HPIV3 genome was identified and it was shown that, in the absence of full genome data, this region could be used for epidemiological surveillance of HPIV3.
Keywords: circulating strains; epidemiology; human parainfluenza 3; phylogenetics.
Conflict of interest statement
No competing interests were disclosed.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

