Investigation into the underlying molecular mechanisms of white adipose tissue through comparative transcriptome analysis of multiple tissues
- PMID: 30569103
- PMCID: PMC6323223
- DOI: 10.3892/mmr.2018.9740
Investigation into the underlying molecular mechanisms of white adipose tissue through comparative transcriptome analysis of multiple tissues
Abstract
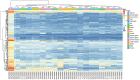
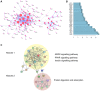
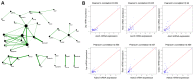
Adipose tissue has a primary role in lipid and glucose metabolism as a storage site for fatty acids, and also functions as an endocrine organ, producing large numbers of hormones and cytokines. Adipose dysfunction triggers a number of obesity‑associated health problems. The aim of the present study was, therefore, to investigate the molecular mechanisms of white adipose tissue (WAT). The GSE9954 microarray data were downloaded from the Gene Expression Omnibus. Adipose‑specific genes were identified through limma package analysis, based on samples of WAT and 17 other types of non‑adipose tissue obtained from mice. Process and pathway enrichment analyses were performed for these genes. Finally, protein‑protein interaction (PPI) and co‑expression networks were constructed and analyzed. In total, 202 adipose‑specific genes were identified, which were involved in key biological processes (including fat cell differentiation and lipid metabolic process) and one key pathway [namely, the adenine monophosphate‑activated protein kinase (AMPK) signaling pathway]. Construction of the PPI network and further molecular complex detection revealed the presence of 17 key genes, including acetyl‑CoA carboxylase α, peroxisome proliferator‑activated receptor (PPAR) γ and leptin, that were involved in the AMPK, PPAR and insulin signaling pathways. In addition, amine oxidase copper containing 3 and adrenoceptor beta 3 were communication hubs in the co‑expression network of adipose‑specific genes. In conclusion, the present study promotes our understanding of the underlying molecular mechanisms of WAT, and may offer an insight into the prevention and treatment of obesity‑associated diseases caused by adipose dysfunction.
Keywords: white adipose tissue; Gene Expression Omnibus; microarray data; differentially expressed genes.
Figures

Similar articles
-
Tissue-specific effects of central leptin on the expression of genes involved in lipid metabolism in liver and white adipose tissue.Endocrinology. 2007 Dec;148(12):5604-10. doi: 10.1210/en.2007-0933. Epub 2007 Sep 6. Endocrinology. 2007. PMID: 17823267
-
Liraglutide regulates lipid metabolism via FGF21- LKB1- AMPK- ACC1 pathway in white adipose tissues and macrophage of type 2 diabetic mice.Biochem Biophys Res Commun. 2021 Apr 9;548:120-126. doi: 10.1016/j.bbrc.2021.02.065. Epub 2021 Feb 25. Biochem Biophys Res Commun. 2021. PMID: 33640604
-
Nr4a1 is required for fasting-induced down-regulation of Pparγ2 in white adipose tissue.Mol Endocrinol. 2013 Jan;27(1):135-49. doi: 10.1210/me.2012-1248. Epub 2012 Dec 18. Mol Endocrinol. 2013. PMID: 23250487 Free PMC article.
-
Adiponectin receptor agonist AdipoRon suppresses adipogenesis in C3H10T1/2 cells through the adenosine monophosphate‑activated protein kinase signaling pathway.Mol Med Rep. 2017 Nov;16(5):7163-7169. doi: 10.3892/mmr.2017.7450. Epub 2017 Sep 8. Mol Med Rep. 2017. PMID: 28901521
-
There and Back Again: Leptin Actions in White Adipose Tissue.Int J Mol Sci. 2020 Aug 21;21(17):6039. doi: 10.3390/ijms21176039. Int J Mol Sci. 2020. PMID: 32839413 Free PMC article. Review.
Cited by
-
Adipokines in Sleep Disturbance and Metabolic Dysfunction: Insights from Network Analysis.Clocks Sleep. 2022 Jun 22;4(3):321-331. doi: 10.3390/clockssleep4030027. Clocks Sleep. 2022. PMID: 35892989 Free PMC article.
-
Obesity of mice lacking VAP-1/SSAO by Aoc3 gene deletion is reproduced in mice expressing a mutated vascular adhesion protein-1 (VAP-1) devoid of amine oxidase activity.J Physiol Biochem. 2021 Feb;77(1):141-154. doi: 10.1007/s13105-020-00756-y. Epub 2020 Jul 25. J Physiol Biochem. 2021. PMID: 32712883
-
Network Pharmacology-Based Analysis and Experimental Exploration of Antidiabetic Mechanisms of Gegen Qinlian Decoction.Front Pharmacol. 2021 Jul 26;12:649606. doi: 10.3389/fphar.2021.649606. eCollection 2021. Front Pharmacol. 2021. PMID: 34381354 Free PMC article.
References
-
- World Health Organization. http://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight. [Mar 8;2018 ];Obesity and overweight.
-
- Moreno-Navarrete JM, Fernández-Real JM. Adipose tissue biology. ‘Vol’. Springer; 2017. Adipocyte differentiation; pp. 69–90. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

