High-Throughput Functional Evaluation of KCNQ1 Decrypts Variants of Unknown Significance
- PMID: 30571187
- PMCID: PMC6309341
- DOI: 10.1161/CIRCGEN.118.002345
High-Throughput Functional Evaluation of KCNQ1 Decrypts Variants of Unknown Significance
Abstract
Background: The explosive growth in known human gene variation presents enormous challenges to current approaches for variant classification that have implications for diagnosis and treatment of many genetic diseases. For disorders caused by mutations in cardiac ion channels as in congenital arrhythmia syndromes, in vitro electrophysiological evidence has high value in discriminating pathogenic from benign variants, but these data are often lacking because assays are cost, time, and labor intensive.
Methods: We implemented a strategy for performing high-throughput functional evaluations of ion channel variants that repurposed an automated electrophysiological recording platform developed previously for drug discovery.
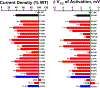
Results: We demonstrated the success of this approach by evaluating 78 variants in KCNQ1, a major gene involved in genetic disorders of cardiac arrhythmia susceptibility. We benchmarked our results with traditional electrophysiological approaches and observed a high level of concordance. This strategy also enabled studies of dominant-negative behavior of variants exhibiting severe loss-of-function. Overall, our results provided functional data useful for reclassifying >65% of the studied KCNQ1 variants.
Conclusions: Our results illustrate an efficient and high-throughput paradigm linking genotype to function for a human cardiac ion channel that will enable data-driven classification of large numbers of variants and create new opportunities for precision medicine.
Keywords: electrophysiology; ion channels; long QT syndrome; mutation; potassium channels.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

