Atg2, Atg9 and Atg18 in mitochondrial integrity, cardiac function and healthspan in Drosophila
- PMID: 30571977
- PMCID: PMC6533900
- DOI: 10.1016/j.yjmcc.2018.12.006
Atg2, Atg9 and Atg18 in mitochondrial integrity, cardiac function and healthspan in Drosophila
Abstract
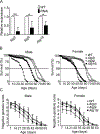
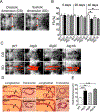
In yeast, the Atg2-Atg18 complex regulates Atg9 recycling from phagophore assembly site during autophagy; their function in higher eukaryotes remains largely unknown. In a targeted screening in Drosophila melanogaster, we show that Mef2-GAL4-RNAi-mediated knockdown of Atg2, Atg9 or Atg18 in the heart and indirect flight muscles led to shortened healthspan (declined locomotive function) and lifespan. These flies displayed an accelerated age-dependent loss of cardiac function along with cardiac hypertrophy (increased heart tube wall thickness) and structural abnormality (distortion of the lumen surface). Using the Mef2-GAL4-MitoTimer mitochondrial reporter system and transmission electron microscopy, we observed significant elongation of mitochondria and reduced number of lysosome-targeted autophagosomes containing mitochondria in the heart tube but exaggerated mitochondrial fragmentation and reduced mitochondrial density in indirect flight muscles. These findings provide the first direct evidence of the importance of Atg2-Atg18/Atg9 autophagy complex in the maintenance of mitochondrial integrity and, regulation of heart and muscle functions in Drosophila, raising the possibility of augmenting Atg2-Atg18/Atg9 activity in promoting mitochondrial health and, muscle and heart function.
Keywords: Autophagy related genes; Cardiac function; Functional aging; Healthspan; Indirect flight muscle; Lifespan; Mitophagy; Negative geotaxis.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of Interest
None
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

