Diagnosis of Bacterial Pathogens in the Urine of Urinary-Tract-Infection Patients Using Surface-Enhanced Raman Spectroscopy
- PMID: 30572659
- PMCID: PMC6321215
- DOI: 10.3390/molecules23123374
Diagnosis of Bacterial Pathogens in the Urine of Urinary-Tract-Infection Patients Using Surface-Enhanced Raman Spectroscopy
Abstract
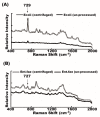
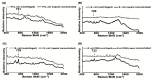
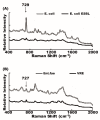
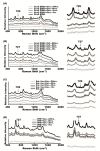
(1) Background: surface-enhanced Raman spectroscopy (SERS) is a novel method for bacteria identification. However, reported applications of SERS in clinical diagnosis are limited. In this study, we used cylindrical SERS chips to detect urine pathogens in urinary tract infection (UTI) patients. (2) Methods: Urine samples were retrieved from 108 UTI patients. A 10 mL urine sample was sent to conventional bacterial culture as a reference. Another 10 mL urine sample was loaded on a SERS chip for bacteria identification and antibiotic susceptibility. We concentrated the urine specimen if the intensity of the Raman spectrum required enhancement. The resulting Raman spectrum was analyzed by a recognition software to compare with spectrum-form reference bacteria and was further confirmed by principal component analysis (PCA). (3) Results: There were 97 samples with single bacteria species identified by conventional urine culture and, among them, 93 can be successfully identified by using SERS without sample concentration. There were four samples that needed concentration for bacteria identification. Antibiotic susceptibility can also be found by SERS. There were seven mixed flora infections found by conventional culture, which can only be identified by the PCA method. (4) Conclusions: SERS can be used in the diagnosis of urinary tract infection with the aid of the recognition software and PCA.
Keywords: Raman spectroscopy; SERS; urinary tract infection.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- Sahm D.F., Thornsberry C., Mayfield D.C., Jones M.E., Karlowsky J.A. Multidrug-resistant urinary tract isolates of Escherichia coli: Prevalence and patient demographics in the United States in 2000. Antimicrob. Agents Chemother. 2001;45:1402–1406. doi: 10.1128/AAC.45.5.1402-1406.2001. - DOI - PMC - PubMed
-
- Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F., Fournier P.E., Drancourt M., La Scola B., Raoult D. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: Impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

