Mechanisms regulating zygotic genome activation
- PMID: 30573849
- PMCID: PMC6558659
- DOI: 10.1038/s41576-018-0087-x
Mechanisms regulating zygotic genome activation
Abstract
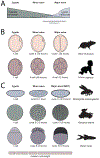
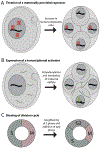
Following fertilization, the two specified gametes must unite to create an entirely new organism. The genome is initially transcriptionally quiescent, allowing the zygote to be reprogrammed into a totipotent state. Gradually, the genome is activated through a process known as the maternal-to-zygotic transition, which enables zygotic gene products to replace the maternal supply that initiated development. This essential transition has been broadly characterized through decades of research in several model organisms. However, we still lack a full mechanistic understanding of how genome activation is executed and how this activation relates to the reprogramming of the zygotic chromatin architecture. Recent work highlights the central role of transcriptional activators and suggests that these factors may coordinate transcriptional activation with other developmental changes.
Figures


References
-
- Gurdon JB The Developmental Capacity of Nuclei taken from Intestinal Epithelium Cells of Feeding Tadpoles. Development 10, (1962). - PubMed
-
- Campbell KHS, McWhir J, Ritchie WA & Wilmut I Sheep cloned by nuclear transfer from a cultured cell line. Nature 380, 64–66 (1996). - PubMed
-
- Newport J & Kirschner M A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30, 687–696 (1982). - PubMed
-
- Tadros W & Lipshitz HD The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

