MicroRNA-31 Reduces the Motility of Proinflammatory T Helper 1 Lymphocytes
- PMID: 30574141
- PMCID: PMC6291424
- DOI: 10.3389/fimmu.2018.02813
MicroRNA-31 Reduces the Motility of Proinflammatory T Helper 1 Lymphocytes
Abstract
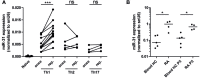
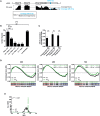
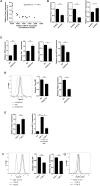
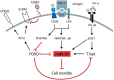
Proinflammatory type 1 T helper (Th1) cells are enriched in inflamed tissues and contribute to the maintenance of chronic inflammation in rheumatic diseases. Here we show that the microRNA- (miR-) 31 is upregulated in murine Th1 cells with a history of repeated reactivation and in memory Th cells isolated from the synovial fluid of patients with rheumatic joint disease. Knock-down of miR-31 resulted in the upregulation of genes associated with cytoskeletal rearrangement and motility and induced the expression of target genes involved in T cell activation, chemokine receptor- and integrin-signaling. Accordingly, inhibition of miR-31 resulted in increased migratory activity of repeatedly activated Th1 cells. The transcription factors T-bet and FOXO1 act as positive and negative regulators of T cell receptor (TCR)-mediated miR-31 expression, respectively. Taken together, our data show that a gene regulatory network involving miR-31, T-bet, and FOXO1 controls the migratory behavior of proinflammatory Th1 cells.
Keywords: CD4; T cell migration; Th1 cells; antagomirs; miR-31; miRNA; regulatory networks; target identification.
Figures


References
-
- Morita Y, Yamamura M, Kawashima M, Harada S, Tsuji K, Shibuya K, et al. . Flow cytometric single-cell analysis of cytokine production by CD4+ T cells in synovial tissue and peripheral blood from patients with rheumatoid arthritis. Arthr Rheum. (1998) 41:1669–76. 10.1002/1529-0131(199809)41:9<1669::AID-ART19>3.0.CO;2-G - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

