VarQ: A Tool for the Structural and Functional Analysis of Human Protein Variants
- PMID: 30574164
- PMCID: PMC6291447
- DOI: 10.3389/fgene.2018.00620
VarQ: A Tool for the Structural and Functional Analysis of Human Protein Variants
Abstract
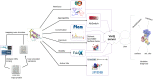
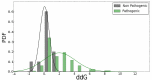
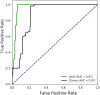
Understanding the functional effect of Single Amino acid Substitutions (SAS), derived from the occurrence of single nucleotide variants (SNVs), and their relation to disease development is a major issue in clinical genomics. Despite the existence of several bioinformatic algorithms and servers that predict if a SAS is pathogenic or not, they give little or no information at all on the reasons for pathogenicity prediction and on the actual predicted effect of the SAS on the protein function. Moreover, few actual methods take into account structural information when available for automated analysis. Moreover, many of these algorithms are able to predict an effect that no necessarily translates directly into pathogenicity. VarQ is a bioinformatic pipeline that incorporates structural information for the detailed analysis and prediction of SAS effect on protein function. It is an online tool which uses UniProt id and automatically analyzes known and user provided SAS for their effect on protein activity, folding, aggregation and protein interactions, among others. We show that structural information, when available, can improve the SAS pathogenicity diagnosis and more important explain its causes. We show that VarQ is able to correctly reproduce previous analysis of RASopathies related mutations, saving extensive and time consuming manual curation. VarQ assessment was performed over a set of previously manually curated RASopathies (diseases that affects the RAS/MAPK signaling pathway) related variants, showing its ability to correctly predict the phenotypic outcome and its underlying cause. This resource is available online at http://varq.qb.fcen.uba.ar/. Supporting Information & Tutorials may be found in the webpage of the tool.
Keywords: FoldX; bioinformatics; single amino acid substitutions; single amino acid substitutions classification; variation diagnosis; web server.
Figures
Similar articles
-
SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants.Nucleic Acids Res. 2012 Jan;40(Database issue):D935-9. doi: 10.1093/nar/gkr996. Epub 2011 Nov 10. Nucleic Acids Res. 2012. PMID: 22075996 Free PMC article.
-
MotSASi: Functional short linear motifs (SLiMs) prediction based on genomic single nucleotide variants and structural data.Biochimie. 2022 Jun;197:59-73. doi: 10.1016/j.biochi.2022.02.002. Epub 2022 Feb 5. Biochimie. 2022. PMID: 35134457
-
Predicting the functional effect of amino acid substitutions and indels.PLoS One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. Epub 2012 Oct 8. PLoS One. 2012. PMID: 23056405 Free PMC article.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
-
ThermoScan: Semi-automatic Identification of Protein Stability Data From PubMed.Front Mol Biosci. 2021 Mar 25;8:620475. doi: 10.3389/fmolb.2021.620475. eCollection 2021. Front Mol Biosci. 2021. PMID: 33842537 Free PMC article. Review.
Cited by
-
The CABANA model 2017-2022: research and training synergy to facilitate bioinformatics applications in Latin America.Front Educ (Lausanne). 2024 Jul 4;9:feduc.2024.1358620. doi: 10.3389/feduc.2024.1358620. Front Educ (Lausanne). 2024. PMID: 39686965 Free PMC article.
-
d-StructMAn: Containerized structural annotation on the scale from genetic variants to whole proteomes.Gigascience. 2022 Sep 20;11:giac086. doi: 10.1093/gigascience/giac086. Gigascience. 2022. PMID: 36130085 Free PMC article.
-
ADDRESS: A Database of Disease-associated Human Variants Incorporating Protein Structure and Folding Stabilities.J Mol Biol. 2021 May 28;433(11):166840. doi: 10.1016/j.jmb.2021.166840. Epub 2021 Feb 2. J Mol Biol. 2021. PMID: 33539887 Free PMC article.
-
Variant Impact Predictor database (VIPdb), version 2: trends from three decades of genetic variant impact predictors.Hum Genomics. 2024 Aug 28;18(1):90. doi: 10.1186/s40246-024-00663-z. Hum Genomics. 2024. PMID: 39198917 Free PMC article.
-
Variant Impact Predictor database (VIPdb), version 2: Trends from 25 years of genetic variant impact predictors.bioRxiv [Preprint]. 2024 Jun 28:2024.06.25.600283. doi: 10.1101/2024.06.25.600283. bioRxiv. 2024. Update in: Hum Genomics. 2024 Aug 28;18(1):90. doi: 10.1186/s40246-024-00663-z. PMID: 38979289 Free PMC article. Updated. Preprint.
References
-
- DeLano L. W. (2002). The PyMOL Molecular Graphics System. Available at: http://pymol.org
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

