Antimicrobial Chemicals Associate with Microbial Function and Antibiotic Resistance Indoors
- PMID: 30574558
- PMCID: PMC6290264
- DOI: 10.1128/mSystems.00200-18
Antimicrobial Chemicals Associate with Microbial Function and Antibiotic Resistance Indoors
Abstract
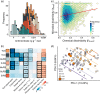
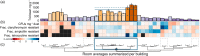
Humans purposefully and inadvertently introduce antimicrobial chemicals into buildings, resulting in widespread compounds, including triclosan, triclocarban, and parabens, in indoor dust. Meanwhile, drug-resistant infections continue to increase, raising concerns that buildings function as reservoirs of, or even select for, resistant microorganisms. Support for these hypotheses is limited largely since data describing relationships between antimicrobials and indoor microbial communities are scant. We combined liquid chromatography-isotope dilution tandem mass spectrometry with metagenomic shotgun sequencing of dust collected from athletic facilities to characterize relationships between indoor antimicrobial chemicals and microbial communities. Elevated levels of triclosan and triclocarban, but not parabens, were associated with distinct indoor microbiomes. Dust of high triclosan content contained increased Gram-positive species with diverse drug resistance capabilities, whose pangenomes were enriched for genes encoding osmotic stress responses, efflux pump regulation, lipid metabolism, and material transport across cell membranes; such triclosan-associated functional shifts have been documented in laboratory cultures but not yet from buildings. Antibiotic-resistant bacterial isolates were cultured from all but one facility, and resistance often increased in buildings with very high triclosan levels, suggesting links between human encounters with viable drug-resistant bacteria and local biocide conditions. This characterization uncovers complex relationships between antimicrobials and indoor microbiomes: some chemicals elicit effects, whereas others may not, and no single functional or resistance factor explained chemical-microbe associations. These results suggest that anthropogenic chemicals impact microbial systems in or around buildings and their occupants, highlighting an emergent need to identify the most important indoor, outdoor, and host-associated sources of antimicrobial chemical-resistome interactions. IMPORTANCE The ubiquitous use of antimicrobial chemicals may have undesired consequences, particularly on microbes in buildings. This study shows that the taxonomy and function of microbes in indoor dust are strongly associated with antimicrobial chemicals-more so than any other feature of the buildings. Moreover, we identified links between antimicrobial chemical concentrations in dust and culturable bacteria that are cross-resistant to three clinically relevant antibiotics. These findings suggest that humans may be influencing the microbial species and genes that are found indoors through the addition and removal of particular antimicrobial chemicals.
Keywords: antibiotic resistance; microbiome; triclosan.
Figures


References
-
- Hartmann EM, Hickey R, Hsu T, Betancourt Román CM, Chen J, Schwager R, Kline J, Brown GZ, Halden RU, Huttenhower C, Green JL. 2016. Antimicrobial chemicals are associated with elevated antibiotic resistance genes in the indoor dust microbiome. Environ Sci Technol 50:9807–9815. doi: 10.1021/acs.est.6b00262. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

