UBC9 Mutant Reveals the Impact of Protein Dynamics on Substrate Selectivity and SUMO Chain Linkages
- PMID: 30574775
- PMCID: PMC6690184
- DOI: 10.1021/acs.biochem.8b01045
UBC9 Mutant Reveals the Impact of Protein Dynamics on Substrate Selectivity and SUMO Chain Linkages
Abstract
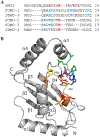
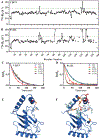
SUMO, a conserved ubiquitin-like protein, is conjugated to a multitude of cellular proteins to maintain genomic integrity and resist genotoxic stress. Studies of the SUMO E2 conjugating enzyme mutant, UBC9P123L, suggested that altered substrate specificity enhances cell sensitivity to DNA damaging agents. Using nuclear magnetic resonance chemical shift studies, we confirm that the mutation does not alter the core globular fold of UBC9, while 15N relaxation measurements demonstrate mutant-induced stabilization of distinct chemical states in residues near the active site cysteine and substrate recognition motifs. We further demonstrate that the P123L substitution induces a switch from the preferential addition of SUMO to lysine residues in unstructured sites to acceptor lysines embedded in secondary structures, thereby also inducing alterations in SUMO chain linkages. Our results provide new insights regarding the impact that structural dynamics of UBC9 have on substrate selection and specifically SUMO chain formation. These findings highlight the potential contribution of nonconsensus SUMO targets and/or alternative SUMO chain linkages on DNA damage response and chemotherapeutic sensitivity.
Figures

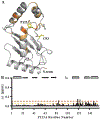




Similar articles
-
Site-specific inhibition of the small ubiquitin-like modifier (SUMO)-conjugating enzyme Ubc9 selectively impairs SUMO chain formation.J Biol Chem. 2017 Sep 15;292(37):15340-15351. doi: 10.1074/jbc.M117.794255. Epub 2017 Aug 7. J Biol Chem. 2017. PMID: 28784659 Free PMC article.
-
Ubc9 sumoylation regulates SUMO target discrimination.Mol Cell. 2008 Aug 8;31(3):371-82. doi: 10.1016/j.molcel.2008.05.022. Mol Cell. 2008. PMID: 18691969
-
Ubc9 sumoylation controls SUMO chain formation and meiotic synapsis in Saccharomyces cerevisiae.Mol Cell. 2013 Jun 6;50(5):625-36. doi: 10.1016/j.molcel.2013.03.027. Epub 2013 May 2. Mol Cell. 2013. PMID: 23644018
-
SUMO: getting it on.Biochem Soc Trans. 2007 Dec;35(Pt 6):1409-13. doi: 10.1042/BST0351409. Biochem Soc Trans. 2007. PMID: 18031233 Review.
-
Enhanced detection of in vivo SUMO conjugation by Ubc9 fusion-dependent sumoylation (UFDS).Methods Mol Biol. 2009;497:63-79. doi: 10.1007/978-1-59745-566-4_5. Methods Mol Biol. 2009. PMID: 19107411 Review.
Cited by
-
The Chronic Toxicity of Endocrine-Disrupting Chemical to Daphnia magna: A Transcriptome and Network Analysis of TNT Exposure.Int J Mol Sci. 2024 Sep 13;25(18):9895. doi: 10.3390/ijms25189895. Int J Mol Sci. 2024. PMID: 39337382 Free PMC article.
-
Topoisomerases and cancer chemotherapy: recent advances and unanswered questions.F1000Res. 2019 Sep 30;8:F1000 Faculty Rev-1704. doi: 10.12688/f1000research.20201.1. eCollection 2019. F1000Res. 2019. PMID: 31602296 Free PMC article. Review.
References
-
- Bergink S; Jentsch S(2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458(7237), 461–7. - PubMed
-
- Seeler J-S; Dejean A(2017) SUMO and the robustness of cancer. Nat Rev Cancer 17(3), 184–197 - PubMed
-
- Flotho A; Melchior F(2013) Sumoylation: A Regulatory Protein Modification in Health and Disease. Annu Rev Biochem 82 (1), 357–385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

