Synthetic mammalian pattern formation driven by differential diffusivity of Nodal and Lefty
- PMID: 30575724
- PMCID: PMC6303393
- DOI: 10.1038/s41467-018-07847-x
Synthetic mammalian pattern formation driven by differential diffusivity of Nodal and Lefty
Abstract
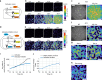
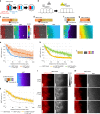
A synthetic mammalian reaction-diffusion pattern has yet to be created, and Nodal-Lefty signaling has been proposed to meet conditions for pattern formation: Nodal is a short-range activator whereas Lefty is a long-range inhibitor. However, this pattern forming possibility has never been directly tested, and the underlying mechanisms of differential diffusivity of Nodal and Lefty remain unclear. Here, through a combination of synthetic and theoretical approaches, we show that a reconstituted Nodal-Lefty network in mammalian cells spontaneously gives rise to a pattern. Surprisingly, extracellular Nodal is confined underneath the cells, resulting in a narrow distribution compared with Lefty. The short-range distribution requires the finger 1 domain of Nodal, and transplantation of the finger 1 domain into Lefty shortens the distribution of Lefty, successfully preventing pattern formation. These results indicate that the differences in localization and domain structures between Nodal and Lefty, combined with the activator-inhibitor topology, are sufficient for reaction-diffusion patterning.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Matsuda, M., Koga, M., Woltjen, K., Nishida, E. & Ebisuya, M. Synthetic lateral inhibition governs cell-type bifurcation with robust ratios. Nat. Commun. 6, 10.1038/ncomms7195 (2015). - PubMed
-
- Solé R, Ollé-Vila A, Vidiella B, Duran-Nebreda S, Conde-Pueyo N. The road to synthetic multicellularity. Curr. Opin. Syst. Biol. 2018;7:60–67. doi: 10.1016/j.coisb.2017.11.007. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

