Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities
- PMID: 30575746
- PMCID: PMC6303322
- DOI: 10.1038/s41467-018-07901-8
Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities
Abstract
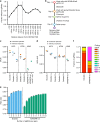
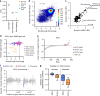
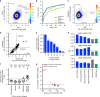
The creation of genome-wide libraries for CRISPR knockout (CRISPRko), interference (CRISPRi), and activation (CRISPRa) has enabled the systematic interrogation of gene function. Here, we show that our recently-described CRISPRko library (Brunello) is more effective than previously published libraries at distinguishing essential and non-essential genes, providing approximately the same perturbation-level performance improvement over GeCKO libraries as GeCKO provided over RNAi. Additionally, we present genome-wide libraries for CRISPRi (Dolcetto) and CRISPRa (Calabrese), and show in negative selection screens that Dolcetto, with fewer sgRNAs per gene, outperforms existing CRISPRi libraries and achieves comparable performance to CRISPRko in detecting essential genes. We also perform positive selection CRISPRa screens and demonstrate that Calabrese outperforms the SAM approach at identifying vemurafenib resistance genes. We further compare CRISPRa to genome-scale libraries of open reading frames (ORFs). Together, these libraries represent a suite of genome-wide tools to efficiently interrogate gene function with multiple modalities.
Conflict of interest statement
J.G.D. consults for Tango Therapeutics. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

