Red fluorescent redox-sensitive biosensor Grx1-roCherry
- PMID: 30576927
- PMCID: PMC6302151
- DOI: 10.1016/j.redox.2018.101071
Red fluorescent redox-sensitive biosensor Grx1-roCherry
Abstract
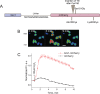
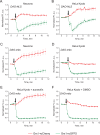
Redox-sensitive fluorescent proteins (roFPs) are a powerful tool for imaging intracellular redox changes. The structure of these proteins contains a pair of cysteines capable of forming a disulfide upon oxidation that affects the protein conformation and spectral characteristics. To date, a palette of such biosensors covers the spectral range from blue to red. However, most of the roFPs suffer from either poor brightness or high pH-dependency, or both. Moreover, there is no roRFP with the redox potential close to that of 2GSH/GSSG redox pair. In the present work, we describe Grx1-roCherry, the first red roFP with canonical FP topology and fluorescent excitation/emission spectra of typical RFP. Grx1-roCherry, with a midpoint redox potential of - 311 mV, is characterized by high brightness and increased pH stability (pKa 6.7). We successfully used Grx1-roCherry in combination with other biosensors in a multiparameter imaging mode to demonstrate redox changes in cells under various metabolic perturbations, including hypoxia/reoxygenation. In particular, using simultaneous expression of Grx1-roCherry and its green analog in various compartments of living cells, we demonstrated that local H2O2 production leads to compartment-specific and cell-type-specific changes in the 2GSH/GSSG ratio. Finally, we demonstrate the utility of Grx1-roCherry for in vivo redox imaging.
Keywords: 2GSH/GSSG; Biosensor; Grx1-roCherry; Multiparameter imaging; Redox-sensitive fluorescent protein.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures




References
-
- Lillig C.H., Berndt C., Holmgren A. Glutaredoxin systems. Biochim. Biophys. Acta. 2008;1780:1304–1317. - PubMed
-
- Fernandes A.P., Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 2004;6:63–74. - PubMed
-
- Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta. 2013;1830:3217–3266. - PubMed
-
- Sedlak J., Lindsay R.H. Estimation of total, protein-bound, and nonprotein sulfhydryl groups in tissue with Ellman's reagent. Anal. Biochem. 1968;25:192–205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

