Regulators of Oncogenic Mutant TP53 Gain of Function
- PMID: 30577483
- PMCID: PMC6356290
- DOI: 10.3390/cancers11010004
Regulators of Oncogenic Mutant TP53 Gain of Function
Abstract
The tumor suppressor p53 (TP53) is the most frequently mutated human gene. Mutations in TP53 not only disrupt its tumor suppressor function, but also endow oncogenic gain-of-function (GOF) activities in a manner independent of wild-type TP53 (wtp53). Mutant TP53 (mutp53) GOF is mainly mediated by its binding with other tumor suppressive or oncogenic proteins. Increasing evidence indicates that stabilization of mutp53 is crucial for its GOF activity. However, little is known about factors that alter mutp53 stability and its oncogenic GOF activities. In this review article, we primarily summarize key regulators of mutp53 stability/activities, including genotoxic stress, post-translational modifications, ubiquitin ligases, and molecular chaperones, as well as a single nucleotide polymorphism (SNP) and dimer-forming mutations in mutp53.
Keywords: TP53; dimer-forming mutation; gain of function; molecular chaperone; mutant TP53; post-translational modification; single nucleotide polymorphism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
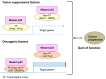
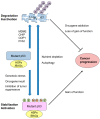
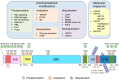
References
-
- Powell B., Soong R., Iacopetta B., Seshadri R., Smith D.R. Prognostic significance of mutations to different structural and functional regions of the p53 gene in breast cancer. Clin. Cancer Res. 2000;6:443–451. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

