Engineering the S-Layer of Caulobacter crescentus as a Foundation for Stable, High-Density, 2D Living Materials
- PMID: 30577690
- PMCID: PMC6945806
- DOI: 10.1021/acssynbio.8b00448
Engineering the S-Layer of Caulobacter crescentus as a Foundation for Stable, High-Density, 2D Living Materials
Abstract
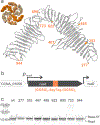
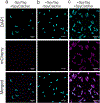
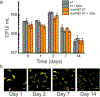
Materials synthesized by organisms, such as bones and wood, combine the ability to self-repair with remarkable mechanical properties. This multifunctionality arises from the presence of living cells within the material and hierarchical assembly of different components across nanometer to micron scales. While creating engineered analogues of these natural materials is of growing interest, our ability to hierarchically order materials using living cells largely relies on engineered 1D protein filaments. Here, we lay the foundation for bottom-up assembly of engineered living material composites in 2D along the cell body using a synthetic biology approach. We engineer the paracrystalline surface-layer (S-layer) of Caulobacter crescentus to display SpyTag peptides that form irreversible isopeptide bonds to SpyCatcher-modified proteins, nanocrystals, and biopolymers on the extracellular surface. Using flow cytometry and confocal microscopy, we show that attachment of these materials to the cell surface is uniform, specific, and covalent, and its density can be controlled on the basis of the insertion location within the S-layer protein, RsaA. Moreover, we leverage the irreversible nature of this attachment to demonstrate via SDS-PAGE that the engineered S-layer can display a high density of materials, reaching 1 attachment site per 288 nm2. Finally, we show that ligation of quantum dots to the cell surface does not impair cell viability, and this composite material remains intact over a period of 2 weeks. Taken together, this work provides a platform for self-organization of soft and hard nanomaterials on a cell surface with precise control over 2D density, composition, and stability of the resulting composite, and is a key step toward building hierarchically ordered engineered living materials with emergent properties.
Keywords: Caulobacter; RsaA; biomaterial; engineered living materials; quantum dots.
Figures




Similar articles
-
Cell-surface display of a Pseudomonas aeruginosa strain K pilin peptide within the paracrystalline S-layer of Caulobacter crescentus.Mol Microbiol. 1997 Oct;26(2):277-88. doi: 10.1046/j.1365-2958.1997.5711932.x. Mol Microbiol. 1997. PMID: 9383153
-
Linker mutagenesis of the Caulobacter crescentus S-layer protein: toward a definition of an N-terminal anchoring region and a C-terminal secretion signal and the potential for heterologous protein secretion.J Bacteriol. 1997 Feb;179(3):601-11. doi: 10.1128/jb.179.3.601-611.1997. J Bacteriol. 1997. PMID: 9006010 Free PMC article.
-
Characterization of mutants of Caulobacter crescentus defective in surface attachment of the paracrystalline surface layer.J Bacteriol. 1994 Oct;176(20):6312-23. doi: 10.1128/jb.176.20.6312-6323.1994. J Bacteriol. 1994. PMID: 7929003 Free PMC article.
-
Cell cycle regulation in Caulobacter: location, location, location.J Cell Sci. 2007 Oct 15;120(Pt 20):3501-7. doi: 10.1242/jcs.005967. J Cell Sci. 2007. PMID: 17928306 Review.
-
Protein localization during the Caulobacter crescentus cell cycle.Curr Opin Microbiol. 1998 Dec;1(6):636-42. doi: 10.1016/s1369-5274(98)80108-2. Curr Opin Microbiol. 1998. PMID: 10066543 Review.
Cited by
-
Resilient living materials built by printing bacterial spores.Nat Chem Biol. 2020 Feb;16(2):126-133. doi: 10.1038/s41589-019-0412-5. Epub 2019 Dec 2. Nat Chem Biol. 2020. PMID: 31792444
-
ALS-ENABLE: creating synergy and opportunity at the Advanced Light Source synchrotron structural biology beamlines.J Synchrotron Radiat. 2025 Jul 1;32(Pt 4):1059-1067. doi: 10.1107/S1600577525004205. Epub 2025 Jun 18. J Synchrotron Radiat. 2025. PMID: 40531663 Free PMC article.
-
Complete atomic structure of a native archaeal cell surface.Cell Rep. 2021 Nov 23;37(8):110052. doi: 10.1016/j.celrep.2021.110052. Cell Rep. 2021. PMID: 34818541 Free PMC article.
-
Robust Self-Regeneratable Stiff Living Materials Fabricated from Microbial Cells.Adv Funct Mater. 2021 May 10;31(19):2010784. doi: 10.1002/adfm.202010784. Epub 2021 Mar 4. Adv Funct Mater. 2021. PMID: 33994904 Free PMC article.
-
A de novo matrix for macroscopic living materials from bacteria.Nat Commun. 2022 Sep 21;13(1):5544. doi: 10.1038/s41467-022-33191-2. Nat Commun. 2022. PMID: 36130968 Free PMC article.
References
-
- Mann S (2001) Biomineralization: Principles and Concepts in Bioinorganic Materials Chemistry, Oxford University Press.

