Classification, substrate specificity and structural features of D-2-hydroxyacid dehydrogenases: 2HADH knowledgebase
- PMID: 30577795
- PMCID: PMC6303947
- DOI: 10.1186/s12862-018-1309-8
Classification, substrate specificity and structural features of D-2-hydroxyacid dehydrogenases: 2HADH knowledgebase
Erratum in
-
Correction to: Classification, substrate specificity and structural features of D-2-hydroxyacid dehydrogenases: 2HADH knowledgebase.BMC Psychiatry. 2019 Jul 16;19(1):221. doi: 10.1186/s12888-019-2207-3. BMC Psychiatry. 2019. PMID: 31311510 Free PMC article.
Abstract
Background: The family of D-isomer specific 2-hydroxyacid dehydrogenases (2HADHs) contains a wide range of oxidoreductases with various metabolic roles as well as biotechnological applications. Despite a vast amount of biochemical and structural data for various representatives of the family, the long and complex evolution and broad sequence diversity hinder functional annotations for uncharacterized members.
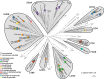
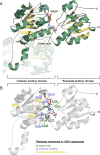
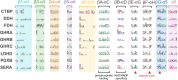
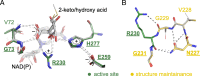
Results: We report an in-depth phylogenetic analysis, followed by mapping of available biochemical and structural data on the reconstructed phylogenetic tree. The analysis suggests that some subfamilies comprising enzymes with similar yet broad substrate specificity profiles diverged early in the evolution of 2HADHs. Based on the phylogenetic tree, we present a revised classification of the family that comprises 22 subfamilies, including 13 new subfamilies not studied biochemically. We summarize characteristics of the nine biochemically studied subfamilies by aggregating all available sequence, biochemical, and structural data, providing comprehensive descriptions of the active site, cofactor-binding residues, and potential roles of specific structural regions in substrate recognition. In addition, we concisely present our analysis as an online 2HADH enzymes knowledgebase.
Conclusions: The knowledgebase enables navigation over the 2HADHs classification, search through collected data, and functional predictions of uncharacterized 2HADHs. Future characterization of the new subfamilies may result in discoveries of enzymes with novel metabolic roles and with properties beneficial for biotechnological applications.
Keywords: D-isomer specific 2-hydroxyacid dehydrogenases; Molecular evolution; Sequence-structure-function relationship; Substrate promiscuity; Substrate specificity.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Stoll VS, Manohar AV, Gillon W, MacFarlane EL, Hynes RC, Pai EF. A thioredoxin fusion protein of VanH, a D-lactate dehydrogenase from enterococcus faecium: cloning, expression, purification, kinetic analysis, and crystallization. Protein Sci. 1998;7(5):1147–1155. doi: 10.1002/pro.5560070508. - DOI - PMC - PubMed
-
- Tarmy E, Kaplan N. Kinetics of Escherichia coli B D-lactate dehydroge and evidence for pyruvate-controlled change in conformation. J Biol Chem. 1968;243(10):2587–2596. - PubMed
-
- Fauvart M, Braeken K, Daniels R, Vos K, Ndayizeye M, Noben JP, Robben J, Vanderleyden J, Michiels J. Identification of a novel glyoxylate reductase supports phylogeny-based enzymatic substrate specificity prediction. Biochim Biophys Acta. 2007;1774(9):1092–1098. doi: 10.1016/j.bbapap.2007.06.009. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

