The interactions of novel mononuclear platinum-based complexes with DNA
- PMID: 30577821
- PMCID: PMC6303901
- DOI: 10.1186/s12885-018-5194-8
The interactions of novel mononuclear platinum-based complexes with DNA
Abstract
Background: Cisplatin has been widely used for the treatment of cancer and its antitumour activity is attributed to its capacity to form DNA adducts, predominantly at guanine residues, which impede cellular processes such as DNA replication and transcription. However, there are associated toxicity and drug resistance issues which plague its use. This has prompted the development and screening of a range of chemotherapeutic drug analogues towards improved efficacy. The biological properties of three novel platinum-based compounds consisting of varying cis-configured ligand groups, as well as a commercially supplied compound, were characterised in this study to determine their potential as anticancer agents.
Methods: The linear amplification reaction was employed, in conjunction with capillary electrophoresis, to quantify the sequence specificity of DNA adducts induced by these compounds using a DNA template containing telomeric repeat sequences. Additionally, the DNA interstrand cross-linking and unwinding efficiency of these compounds were assessed through the application of denaturing and native agarose gel electrophoresis techniques, respectively. Their cytotoxicity was determined in HeLa cells using a colorimetric cell viability assay.
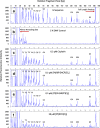
Results: All three novel platinum-based compounds were found to induce DNA adduct formation at the tandem telomeric repeat sequences. The sequence specificity profile at these sites was characterised and these were distinct from that of cisplatin. Two of these compounds with the enantiomeric 1,2-diaminocyclopentane ligand (SS and RR-DACP) were found to induce a greater degree of DNA unwinding than cisplatin, but exhibited marginally lower DNA cross-linking efficiencies. Furthermore, the RR-isomer was more cytotoxic in HeLa cells than cisplatin.
Conclusions: The biological characteristics of these compounds were assessed relative to cisplatin, and a variation in the sequence specificity and a greater capacity to induce DNA unwinding was observed. These compounds warrant further investigations towards developing more efficient chemotherapeutic drugs.
Keywords: Anticancer drug; Cisplatin; DNA adducts; Interstrand cross-linking; Linear amplification reaction; Sequence specificity; Telomeric repeat.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable. However, we wish to acknowledge that in 1951 HeLa cells were taken without consent from Henrietta Lacks and we thank the Lacks family for their generous contributions to the biomedical community.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

