Conformational flexibility in the enterovirus RNA replication platform
- PMID: 30578285
- PMCID: PMC6380274
- DOI: 10.1261/rna.069476.118
Conformational flexibility in the enterovirus RNA replication platform
Abstract
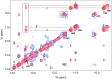
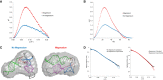
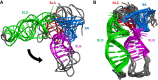
A presumed RNA cloverleaf (5'CL), located at the 5'-most end of the noncoding region of the enterovirus genome, is the primary established site for initiation of genomic replication. Stem-loop B (SLB) and stem-loop D (SLD), the two largest stem-loops within the 5'CL, serve as recognition sites for protein interactions that are essential for replication. Here we present the solution structure of rhinovirus serotype 14 5'CL using a combination of nuclear magnetic resonance spectroscopy and small-angle X-ray scattering. In the absence of magnesium, the structure adopts an open, somewhat extended conformation. In the presence of magnesium, the structure compacts, bringing SLB and SLD into close contact, a geometry that creates an extensive accessible major groove surface, and permits interaction between the proteins that target each stem-loop.
Keywords: NMR; RNA; SAXS; cloverleaf; enterovirus; replication.
© 2019 Warden et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





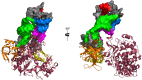
References
Publication types
MeSH terms
Substances
Grants and funding
- S10RR025062 /NH/NIH HHS/United States
- S10 RR025062/RR/NCRR NIH HHS/United States
- S10RR029220 /NH/NIH HHS/United States
- S10 RR029220/RR/NCRR NIH HHS/United States
- P41GM103399 /NH/NIH HHS/United States
- S10RR08438 /NH/NIH HHS/United States
- S10RR023438 /NH/NIH HHS/United States
- S10RR02781 /NH/NIH HHS/United States
- R35 GM118131/GM/NIGMS NIH HHS/United States
- R35GM118131 /NH/NIH HHS/United States
- S10 RR002781/RR/NCRR NIH HHS/United States
- P41 GM103399/GM/NIGMS NIH HHS/United States
- S10 RR008438/RR/NCRR NIH HHS/United States
- S10 RR023438/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
