TICA: Transcriptional Interaction and Coregulation Analyzer
- PMID: 30578913
- PMCID: PMC6364043
- DOI: 10.1016/j.gpb.2018.05.004
TICA: Transcriptional Interaction and Coregulation Analyzer
Abstract
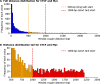
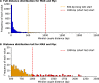
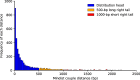
Transcriptional regulation is critical to cellular processes of all organisms. Regulatory mechanisms often involve more than one transcription factor (TF) from different families, binding together and attaching to the DNA as a single complex. However, only a fraction of the regulatory partners of each TF is currently known. In this paper, we present the Transcriptional Interaction and Coregulation Analyzer (TICA), a novel methodology for predicting heterotypic physical interaction of TFs. TICA employs a data-driven approach to infer interaction phenomena from chromatin immunoprecipitation and sequencing (ChIP-seq) data. Its prediction rules are based on the distribution of minimal distance couples of paired binding sites belonging to different TFs which are located closest to each other in promoter regions. Notably, TICA uses only binding site information from input ChIP-seq experiments, bypassing the need to do motif calling on sequencing data. We present our method and test it on ENCODE ChIP-seq datasets, using three cell lines as reference including HepG2, GM12878, and K562. TICA positive predictions on ENCODE ChIP-seq data are strongly enriched when compared to protein complex (CORUM) and functional interaction (BioGRID) databases. We also compare TICA against both motif/ChIP-seq based methods for physical TF-TF interaction prediction and published literature. Based on our results, TICA offers significant specificity (average 0.902) while maintaining a good recall (average 0.284) with respect to CORUM, providing a novel technique for fast analysis of regulatory effect in cell lines. Furthermore, predictions by TICA are complementary to other methods for TF-TF interaction prediction (in particular, TACO and CENTDIST). Thus, combined application of these prediction tools results in much improved sensitivity in detecting TF-TF interactions compared to TICA alone (sensitivity of 0.526 when combining TICA with TACO and 0.585 when combining with CENTDIST) with little compromise in specificity (specificity 0.760 when combining with TACO and 0.643 with CENTDIST). TICA is publicly available at http://geco.deib.polimi.it/tica/.
Keywords: Coregulation; Data-driven analysis; Machine learning; Protein–protein interactions; Transcription factors.
Copyright © 2018 The Authors. Production and hosting by Elsevier B.V. All rights reserved.
Figures


References
-
- Hughes T.R. Springer, Netherlands; Berlin: 2011. A handbook of transcription factors. - PubMed
-
- Weirauch M.T., Hughes T.R. A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. In: Hughes T.R., editor. A handbook of transcription factors. Springer; Netherlands, Berlin: 2011. pp. 26–73. - PubMed
-
- Jolma A., Yin Y., Nitta K.R., Dave K., Popov A., Taipale M. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527:384–389. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

