Fatty Acids Compete with Aβ in Binding to Serum Albumin by Quenching Its Conformational Flexibility
- PMID: 30580919
- PMCID: PMC6349961
- DOI: 10.1016/j.bpj.2018.11.3133
Fatty Acids Compete with Aβ in Binding to Serum Albumin by Quenching Its Conformational Flexibility
Abstract
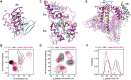
Human serum albumin (HSA) has been identified as an important regulator of amyloid-β (Aβ) fibrillization both in blood plasma and in cerebrospinal fluid. Fatty acids bind to HSA, and high serum levels of fatty acids increase the risk of Alzheimer's disease. In vitro, fatty-acid-loaded HSA (FA·HSA) loses the protective effect against Aβ fibrillization, but the mechanism underlying the interference of fatty acids on Aβ-HSA interactions has been unclear. Here, we used molecular dynamics simulations to gain atomic-level insight on the weak binding of monomeric Aβ40 and Aβ42 peptides with apo and FA·HSA. Consistent with recent NMR data, C-terminal residues of the Aβ peptides have the highest propensities for interacting with apo HSA. Interestingly, the Aβ binding residues of apo and FA·HSA exhibit distinct patterns, which qualitatively correlate with backbone flexibility. In FA·HSA, both flexibilities and Aβ binding propensities are relatively even among the three domains. In contrast, in apo HSA, domain III shows the highest flexibility and is the primary target for Aβ binding. Specifically, deformation of apo HSA creates strong binding sites within subdomain IIIb, around the interface between subdomains IIIa and IIIb, and at the cleft between domains III and I. Therefore, much like disordered proteins, HSA can take advantage of flexibility in forming promiscuous interactions with partners, until the flexibility is quenched by fatty-acid binding. Our work explains the effect of fatty acids on Aβ-HSA binding and contributes to the understanding of HSA regulation of Aβ aggregation.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Kuo Y.M., Emmerling M.R., Roher A.E. High levels of circulating Abeta42 are sequestered by plasma proteins in Alzheimer’s disease. Biochem. Biophys. Res. Commun. 1999;257:787–791. - PubMed
-
- Bohrmann B., Tjernberg L., Nordstedt C. Endogenous proteins controlling amyloid beta-peptide polymerization. Possible implications for beta-amyloid formation in the central nervous system and in peripheral tissues. J. Biol. Chem. 1999;274:15990–15995. - PubMed
-
- Biere A.L., Ostaszewski B., Selkoe D.J. Amyloid βετα-peptide is transported on lipoproteins and albumin in human plasma. J. Biol. Chem. 1996;271:32916–32922. - PubMed
-
- Kuo Y.M., Kokjohn T.A., Roher A.E. Amyloid-βετα peptides interact with plasma proteins and erythrocytes: implications for their quantitation in plasma. Biochem. Biophys. Res. Commun. 2000;268:750–756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

