p53 Represses the Mevalonate Pathway to Mediate Tumor Suppression
- PMID: 30580964
- PMCID: PMC6483089
- DOI: 10.1016/j.cell.2018.11.011
p53 Represses the Mevalonate Pathway to Mediate Tumor Suppression
Abstract
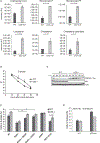
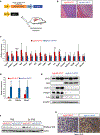
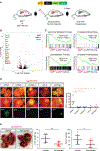
There are still gaps in our understanding of the complex processes by which p53 suppresses tumorigenesis. Here we describe a novel role for p53 in suppressing the mevalonate pathway, which is responsible for biosynthesis of cholesterol and nonsterol isoprenoids. p53 blocks activation of SREBP-2, the master transcriptional regulator of this pathway, by transcriptionally inducing the ABCA1 cholesterol transporter gene. A mouse model of liver cancer reveals that downregulation of mevalonate pathway gene expression by p53 occurs in premalignant hepatocytes, when p53 is needed to actively suppress tumorigenesis. Furthermore, pharmacological or RNAi inhibition of the mevalonate pathway restricts the development of murine hepatocellular carcinomas driven by p53 loss. Like p53 loss, ablation of ABCA1 promotes murine liver tumorigenesis and is associated with increased SREBP-2 maturation. Our findings demonstrate that repression of the mevalonate pathway is a crucial component of p53-mediated liver tumor suppression and outline the mechanism by which this occurs.
Keywords: ABCA1; SREBP-2; cancer metabolism; mevalonate pathway; p53; tumor suppression.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
C.P. is a member of the scientific advisory board of Aileron Therapeutics. S.W.L. is a founder and scientific advisory board member of Mirimus, Inc., Blueprint Medicines, and ORIC Pharmaceuticals. S.W.L. is also a scientific advisory board member of Constellation Pharmaceuticals and PMV Pharmaceuticals.
Figures




References
-
- Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, and Goldstein JL (2004). Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem 279, 52772–52780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

