A Diet-Sensitive Commensal Lactobacillus Strain Mediates TLR7-Dependent Systemic Autoimmunity
- PMID: 30581114
- PMCID: PMC6377154
- DOI: 10.1016/j.chom.2018.11.009
A Diet-Sensitive Commensal Lactobacillus Strain Mediates TLR7-Dependent Systemic Autoimmunity
Abstract
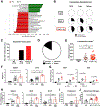
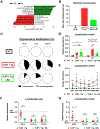
Western lifestyle is linked to autoimmune and metabolic diseases, driven by changes in diet and gut microbiota composition. Using Toll-like receptor 7 (TLR7)-dependent mouse models of systemic lupus erythematosus (SLE), we dissect dietary effects on the gut microbiota and find that Lactobacillus reuteri can drive autoimmunity but is ameliorated by dietary resistant starch (RS). Culture of internal organs and 16S rDNA sequencing revealed TLR7-dependent translocation of L. reuteri in mice and fecal enrichment of Lactobacillus in a subset of SLE patients. L. reuteri colonization worsened autoimmune manifestations under specific-pathogen-free and gnotobiotic conditions, notably increasing plasmacytoid dendritic cells (pDCs) and interferon signaling. However, RS suppressed the abundance and translocation of L. reuteri via short-chain fatty acids, which inhibited its growth. Additionally, RS decreased pDCs, interferon pathways, organ involvement, and mortality. Thus, RS exerts beneficial effects in lupus-prone hosts through suppressing a pathobiont that promotes interferon pathways implicated in the pathogenesis of human autoimmunity.
Keywords: Clostridiaceae; Clostridiales; Lactobacillus; Lactobacillus reuteri; gut microbiota; human SLE microbiome; interferon; plasmacytoid dendritic cells; resistant starch; systemic lupus erythematosus.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests
M.A.K. received salary, consulting fees, honoraria and research funds from Roche, Bristol-Meyers Squibb, AbbVie, and Cell Applications.
Figures





Comment in
-
Dietary modulation of the microbiome as therapy.Nat Rev Rheumatol. 2019 Feb;15(2):66. doi: 10.1038/s41584-019-0165-2. Nat Rev Rheumatol. 2019. PMID: 30622298 No abstract available.
-
Fiber Puts Lactobacillus to SLEep.Cell Host Microbe. 2019 Jan 9;25(1):3-5. doi: 10.1016/j.chom.2018.12.012. Cell Host Microbe. 2019. PMID: 30629916
References
-
- Bourn R, and James JA (2016). Pre-Clinical Lupus. Curr. Opin. Rheumatol 8, 444–454.
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis ofhigh- throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat. Methods 7, 335–336. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

