Modulation of faecal metagenome in Crohn's disease: Role of microRNAs as biomarkers
- PMID: 30581271
- PMCID: PMC6295834
- DOI: 10.3748/wjg.v24.i46.5223
Modulation of faecal metagenome in Crohn's disease: Role of microRNAs as biomarkers
Abstract
Background: The gut microbiota plays a key role in the maintenance of intestinal homeostasis and the development and activation of the host immune system. It has been shown that commensal bacterial species can regulate the expression of host genes. 16S rRNA gene sequencing has shown that the microbiota in inflammatory bowel disease (IBD) is abnormal and characterized by reduced diversity. MicroRNAs (miRNAs) have been explored as biomarkers and therapeutic targets, since they are able to regulate specific genes associated with Crohn's disease (CD). In this work, we aim to investigate the composition of gut microbiota of active treatment-naïve adult CD patients, with miRNA profile from gut microbiota.
Aim: To investigate the composition of gut microbiota of active treatment-naïve adult CD patients, with miRNA profile from gut microbiota.
Methods: Patients attending the outpatient clinics at Valme University Hospital without relevant co-morbidities were matched according to age and gender. Faecal samples of new-onset CD patients, free of treatment, and healthy controls were collected. Faecal samples were homogenized, and DNA was amplified by PCR using primers directed to the 16S bacterial rRNA gene. Pyrosequencing was performed using GS-Junior platform. For sequence analysis, MG-RAST server with the database Ribosomal Project was used. MiRNA profile and their relative abundance were analyzed by quantitative PCR.
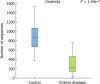
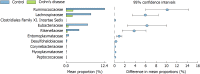
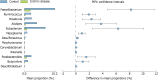
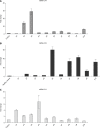
Results: Microbial community was characterized using 16S rRNA gene sequencing in 29 samples (n = 13 CD patients, and n = 16 healthy controls). The mean Shannon diversity was higher in the healthy control population compared to CD group (5.5 vs 3.7). A reduction in Firmicutes and an increase in Bacteroidetes were found. Clostridia class was also significantly reduced in CD. Principal components analysis showed a grouping pattern, identified in most of the subjects in both groups, showing a marked difference between control and CD groups. A functional metabolic study showed that a lower metabolism of carbohydrates (P = 0.000) was found in CD group, while the metabolism of lipids was increased. In CD patients, three miRNAs were induced in affected mucosa: mir-144 (6.2 ± 1.3 fold), mir-519 (21.8 ± 3.1) and mir-211 (2.3 ± 0.4).
Conclusion: Changes in microbial function in active non-treated CD subjects and three miRNAs in affected vs non-affected mucosa have been found. miRNAs profile may serve as a biomarker.
Keywords: Bacteroidetes; Crohn’s disease; Dysbiosis; Firmicutes; microRNAs.
Conflict of interest statement
Conflict-of-interest statement: All authors declare there are no competing interests in this study.
Figures


References
-
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. - PubMed

