Distinct Nasopharyngeal and Oropharyngeal Microbiota of Children with Influenza A Virus Compared with Healthy Children
- PMID: 30581863
- PMCID: PMC6276510
- DOI: 10.1155/2018/6362716
Distinct Nasopharyngeal and Oropharyngeal Microbiota of Children with Influenza A Virus Compared with Healthy Children
Abstract
Background: Influenza A virus (IAV) has had the highest morbidity globally over the past decade. A growing number of studies indicate that the upper respiratory tract (URT) microbiota plays a key role for respiratory health and that a dysfunctional respiratory microbiota is associated with disease; but the impact of microbiota during influenza is understudied.
Methods: We recruited 180 children, including 121 IAV patients and 59 age-matched healthy children. Nasopharyngeal (NP) and oropharyngeal (OP) swabs were collected to conduct 16S rDNA sequencing and compare microbiota structures in different individuals.
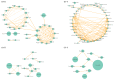
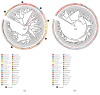
Results: Both NP and OP microbiota in IAV patients differed from those in healthy individuals. The NP dominated genera in IVA patients, such as Moraxella, Staphylococcus, Corynebacterium, and Dolosigranulum, showed lower abundance than in healthy children. The Streptococcus significantly enriched in patients' NP and Phyllobacterium could be generally detected in patients' NP microbiota. The most abundant genera in OP microbiota showed a decline tendency in patients, including Streptococcus, Neisseria, and Haemophilus. The URT's bacterial concurrence network changed dramatically in patients. NP and OP samples were clustered into subgroups by different dominant genera; and NP and OP microbiota provided the precise indicators to distinguish IAV patients from healthy children.
Conclusion: This is the first respiratory microbiome analysis on pediatric IAV infection which reveals distinct NP and OP microbiota in influenza patients. It provides a new insight into IAV research from the microecology aspect and promotes the understanding of IAV pathogenesis.
Figures


References
-
- Bradley J. S., Byington C. L., Shah S. S., et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: Clinical practice guidelines by the pediatric infectious diseases society and the infectious diseases society of America. Clinical Infectious Diseases. 2011;53(7):e25–e76. doi: 10.1093/cid/cir531. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

