Mechanisms of opening and closing of the bacterial replicative helicase
- PMID: 30582519
- PMCID: PMC6391071
- DOI: 10.7554/eLife.41140
Mechanisms of opening and closing of the bacterial replicative helicase
Abstract
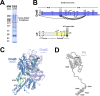
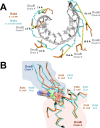
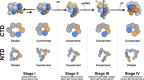
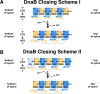
Assembly of bacterial ring-shaped hexameric replicative helicases on single-stranded (ss) DNA requires specialized loading factors. However, mechanisms implemented by these factors during opening and closing of the helicase, which enable and restrict access to an internal chamber, are not known. Here, we investigate these mechanisms in the Escherichia coli DnaB helicase•bacteriophage λ helicase loader (λP) complex. We show that five copies of λP bind at DnaB subunit interfaces and reconfigure the helicase into an open spiral conformation that is intermediate to previously observed closed ring and closed spiral forms; reconfiguration also produces openings large enough to admit ssDNA into the inner chamber. The helicase is also observed in a restrained inactive configuration that poises it to close on activating signal, and transition to the translocation state. Our findings provide insights into helicase opening, delivery to the origin and ssDNA entry, and closing in preparation for translocation.
Keywords: DNA replication; DnaB replicative helicase; E. coli; biochemistry; chemical biology; cryogenic electron microscopy; helicase loader; molecular biophysics; replication initiation; structural biology.
© 2018, Chase et al.
Conflict of interest statement
JC, AC, AN, EE, PO, KM, DP, MS, BC, Ad, DJ No competing interests declared
Figures












Similar articles
-
Convergent evolution in two bacterial replicative helicase loaders.Trends Biochem Sci. 2022 Jul;47(7):620-630. doi: 10.1016/j.tibs.2022.02.005. Epub 2022 Mar 26. Trends Biochem Sci. 2022. PMID: 35351361 Free PMC article. Review.
-
Cryptic single-stranded-DNA binding activities of the phage lambda P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA.Proc Natl Acad Sci U S A. 1997 Feb 18;94(4):1154-9. doi: 10.1073/pnas.94.4.1154. Proc Natl Acad Sci U S A. 1997. PMID: 9037022 Free PMC article.
-
Structural insight into replicative helicase loading in Escherichia coli.J Biochem. 2022 May 27;171(6):605-607. doi: 10.1093/jb/mvac023. J Biochem. 2022. PMID: 35238386
-
Distinct Quaternary States, Intermediates, and Autoinhibition During Loading of the DnaB-Replicative Helicase by the Phage λP Helicase Loader.bioRxiv [Preprint]. 2025 Jun 2:2022.12.30.522210. doi: 10.1101/2022.12.30.522210. bioRxiv. 2025. PMID: 40501539 Free PMC article. Preprint.
-
Structural Insight Into the Function of DnaB Helicase in Bacterial DNA Replication.Proteins. 2025 Feb;93(2):420-429. doi: 10.1002/prot.26746. Epub 2024 Sep 4. Proteins. 2025. PMID: 39230358 Review.
Cited by
-
Native Mass Spectrometry-Based Screening for Optimal Sample Preparation in Single-Particle Cryo-EM.Structure. 2021 Feb 4;29(2):186-195.e6. doi: 10.1016/j.str.2020.11.001. Epub 2020 Nov 19. Structure. 2021. PMID: 33217329 Free PMC article.
-
Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.Nucleic Acids Res. 2021 Jun 21;49(11):6569-6586. doi: 10.1093/nar/gkab463. Nucleic Acids Res. 2021. PMID: 34107018 Free PMC article.
-
Structural insights into human topoisomerase 3β DNA and RNA catalysis and nucleic acid gate dynamics.Nat Commun. 2025 Jan 19;16(1):834. doi: 10.1038/s41467-025-55959-y. Nat Commun. 2025. PMID: 39828754 Free PMC article.
-
Convergent evolution in two bacterial replicative helicase loaders.Trends Biochem Sci. 2022 Jul;47(7):620-630. doi: 10.1016/j.tibs.2022.02.005. Epub 2022 Mar 26. Trends Biochem Sci. 2022. PMID: 35351361 Free PMC article. Review.
-
The LH-DH module of bacterial replicative helicases is the common binding site for DciA and other helicase loaders.Acta Crystallogr D Struct Biol. 2023 Feb 1;79(Pt 2):177-187. doi: 10.1107/S2059798323000281. Epub 2023 Feb 6. Acta Crystallogr D Struct Biol. 2023. PMID: 36762863 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM128303/GM/NIGMS NIH HHS/United States
- P41 GM109824/NH/NIH HHS/United States
- P41 GM103310/GM/NIGMS NIH HHS/United States
- P41 GM103314/GM/NIGMS NIH HHS/United States
- G12 MD007603/MD/NIMHD NIH HHS/United States
- R01 GM084162/GM/NIGMS NIH HHS/United States
- GM084162/NH/NIH HHS/United States
- S10 OD019994/OD/NIH HHS/United States
- OD019994/NH/NIH HHS/United States
- P41 GM103314/NH/NIH HHS/United States
- F32GM128303/NH/NIH HHS/United States
- 5G12MD007603-30/NH/NIH HHS/United States
- P41 GM109824/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

