SMC5/6: Multifunctional Player in Replication
- PMID: 30583551
- PMCID: PMC6356406
- DOI: 10.3390/genes10010007
SMC5/6: Multifunctional Player in Replication
Abstract
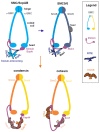
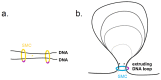
The genome replication process is challenged at many levels. Replication must proceed through different problematic sites and obstacles, some of which can pause or even reverse the replication fork (RF). In addition, replication of DNA within chromosomes must deal with their topological constraints and spatial organization. One of the most important factors organizing DNA into higher-order structures are Structural Maintenance of Chromosome (SMC) complexes. In prokaryotes, SMC complexes ensure proper chromosomal partitioning during replication. In eukaryotes, cohesin and SMC5/6 complexes assist in replication. Interestingly, the SMC5/6 complexes seem to be involved in replication in many ways. They stabilize stalled RFs, restrain RF regression, participate in the restart of collapsed RFs, and buffer topological constraints during RF progression. In this (mini) review, I present an overview of these replication-related functions of SMC5/6.
Keywords: SMC complexes; SMC5/6; chromatin structure; collapsed replication fork; homologous recombination; loop extrusion; replication fork regression; stalled replication fork.
Conflict of interest statement
The author declares no conflicts of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

