Spatial correlations exploitation based on nonlocal voxel-wise GWAS for biomarker detection of AD
- PMID: 30584014
- PMCID: PMC6413305
- DOI: 10.1016/j.nicl.2018.101642
Spatial correlations exploitation based on nonlocal voxel-wise GWAS for biomarker detection of AD
Abstract
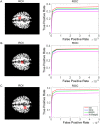
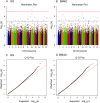
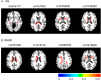
Potential biomarker detection is a crucial area of study for the prediction, diagnosis, and monitoring of Alzheimer's disease (AD). The voxelwise genome-wide association study (vGWAS) is widely used in imaging genomics studies that is usually applied to the detection of AD biomarkers in both imaging and genetic data. However, performing vGWAS remains a challenge because of the computational complexity of the technique and our ignorance of the spatial correlations within the imaging data. In this paper, we propose a novel method based on the exploitation of spatial correlations that may help to detect potential AD biomarkers using a fast vGWAS. To incorporate spatial correlations, we applied a nonlocal method that supposed that a given voxel could be represented by weighting the sum of the other voxels. Three commonly used weighting methods were adopted to calculate the weights among different voxels in this study. Then, a fast vGWAS approach was used to assess the association between the image and the genetic data. The proposed method was estimated using both simulated and real data. In the simulation studies, we designed a set of experiments to evaluate the effectiveness of the nonlocal method for incorporating spatial correlations in vGWAS. The experiments showed that incorporating spatial correlations by the nonlocal method could improve the detecting accuracy of AD biomarkers. For real data, we successfully identified three genes, namely, ANK3, MEIS2, and TLR4, which have significant associations with mental retardation, learning disabilities and age according to previous research. These genes have profound impacts on AD or other neurodegenerative diseases. Our results indicated that our method might be an effective and valuable tool for detecting potential biomarkers of AD.
Keywords: AD biomarker; Imaging genomics studies; Nonlocal method; Spatial correlations; vGWAS.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2004;21(2):263–265. - PubMed
-
- Braskie M.N., Jahanshad N., Stein J.L., Barysheva M., Mcmahon K.L., de Zubicaray G.I., Martin N.G., Wright M.J., Ringman J.M., Toga A.W., Thompson P.M. Common Alzheimer's disease risk variant within the CLU gene affects white matter microstructure in young adults. J. Neurosci. 2011;31(18):6764–6770. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

