Seven Genes Based Novel Signature Predicts Clinical Outcome and Platinum Sensitivity of High Grade IIIc Serous Ovarian Carcinoma
- PMID: 30585265
- PMCID: PMC6299362
- DOI: 10.7150/ijbs.28249
Seven Genes Based Novel Signature Predicts Clinical Outcome and Platinum Sensitivity of High Grade IIIc Serous Ovarian Carcinoma
Abstract
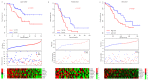
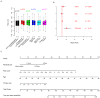
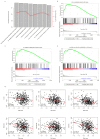
Background: As a major subtype of ovarian cancer, high grade FIGO stage IIIc serous ovarian carcinoma (HG3cSOC), has various prognosis due to genetic heterogeneity. Methods: The transcriptome of 401 primary FIGO IIIc serous ovarian samples was screened, seven genes based prognostic model was developed. The prognostic valueof risk score in four different cohorts (TCGA-cohort, Poland-cohort, Japan-cohort and USA-cohort) was validated. The relationship between risk score and other clinical indicators were analyzed. The guide value of risk score for platinum-taxol chemotherapy was also assayed. Tissue microenvironment difference among samples with different risk scores was investigated. Results: High-risk group (N=200, median survival months: 39.6, 95% CI: 35.9-46.3 months) had a significantly worse prognosis than low-risk group (N=201, median survival months: 52.6, 95% CI: 45.2-64.9 months;). The risk score's performance was validated in Japan-cohort (N=90, Poland-cohort (N=48) and USA-cohort (N=84). The risk score is independent from age, primary tumor size, grade and treatment methods and the performance of risk score is uniform in subgroups. Furthermore, the risk score predicted the response of HG3cSOC to platinum-based regimen after surgery, and this finding was further validated in newly collected China-cohort (N=102). Gene Set Enrichment Analysis (GSEA) and tumor infiltration analysis revealed that risk score reflected the immune infiltration and cell-cell interaction status, and the migration function of candidate genes were also verified. Conclusions: The optimized seven genes-based model is a valuable and robust model in predicting the survival of HG3cSOC, and served as a valuable marker for the response to platinum-based chemotherapy.
Keywords: High grade FIGO IIIc serous ovarian carcinoma; chemotherapy; microenvironment.; model; prognosis; transcriptome.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




Similar articles
-
Transcriptome-based signature predicts the effect of taxol in serous ovarian cancer.PLoS One. 2018 Mar 1;13(3):e0192812. doi: 10.1371/journal.pone.0192812. eCollection 2018. PLoS One. 2018. PMID: 29494610 Free PMC article.
-
HOXA4/HOXB3 gene expression signature as a biomarker of recurrence in patients with high-grade serous ovarian cancer following primary cytoreductive surgery and first-line adjuvant chemotherapy.Gynecol Oncol. 2018 Apr;149(1):155-162. doi: 10.1016/j.ygyno.2018.01.022. Gynecol Oncol. 2018. PMID: 29402501
-
Interferon regulatory factor 1 is an independent predictor of platinum resistance and survival in high-grade serous ovarian carcinoma.Gynecol Oncol. 2014 Sep;134(3):591-8. doi: 10.1016/j.ygyno.2014.06.025. Epub 2014 Jul 1. Gynecol Oncol. 2014. PMID: 24995581
-
Potential Transcriptomic Biomarkers for Predicting Platinum-based Chemotherapy Resistance in Patients With High-grade Serous Ovarian Cancer.Anticancer Res. 2024 Nov;44(11):4691-4707. doi: 10.21873/anticanres.17296. Anticancer Res. 2024. PMID: 39477310 Review.
-
Systematic analysis reveals a lncRNA-mRNA co-expression network associated with platinum resistance in high-grade serous ovarian cancer.Invest New Drugs. 2018 Apr;36(2):187-194. doi: 10.1007/s10637-017-0523-3. Epub 2017 Oct 30. Invest New Drugs. 2018. PMID: 29082457
Cited by
-
PRSS1 Upregulation Predicts Platinum Resistance in Ovarian Cancer Patients.Front Cell Dev Biol. 2021 Jan 28;8:618341. doi: 10.3389/fcell.2020.618341. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33585454 Free PMC article.
-
Development and verification of an immune-related gene pairs prognostic signature in ovarian cancer.J Cell Mol Med. 2021 Mar;25(6):2918-2930. doi: 10.1111/jcmm.16327. Epub 2021 Feb 4. J Cell Mol Med. 2021. PMID: 33543590 Free PMC article.
-
Prognostic alternative splicing signature reveals the landscape of immune infiltration in Pancreatic Cancer.J Cancer. 2020 Sep 21;11(22):6530-6544. doi: 10.7150/jca.47877. eCollection 2020. J Cancer. 2020. PMID: 33046974 Free PMC article.
-
Identification of a glycolysis-related gene signature for survival prediction of ovarian cancer patients.Cancer Med. 2021 Nov;10(22):8222-8237. doi: 10.1002/cam4.4317. Epub 2021 Oct 5. Cancer Med. 2021. PMID: 34609082 Free PMC article.
-
A risk model of gene signatures for predicting platinum response and survival in ovarian cancer.J Ovarian Res. 2022 Mar 31;15(1):39. doi: 10.1186/s13048-022-00969-3. J Ovarian Res. 2022. PMID: 35361267 Free PMC article.
References
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115–32. - PubMed
-
- Bakkar R, Gershenson D, Fox P, Vu K, Zenali M, Silva E. Stage IIIC ovarian/peritoneal serous carcinoma: a heterogeneous group of patients with different prognoses. International journal of gynecological pathology: official journal of the International Society of Gynecological Pathologists. 2014;33:302–8. - PubMed
-
- Yang S, Zhang Y, Meng F, Liu Y, Xia B, Xiao M. et al. Overexpression of multiple myeloma SET domain (MMSET) is associated with advanced tumor aggressiveness and poor prognosis in serous ovarian carcinoma. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2013;18:257–63. - PubMed
-
- Lohneis P, Darb-Esfahani S, Dietel M, Braicu I, Sehouli J, Arsenic R. PDK1 is Expressed in Ovarian Serous Carcinoma and Correlates with Improved Survival in High-grade Tumors. Anticancer research. 2015;35:6329–34. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

