Major contribution of transcription initiation to 5'-end formation of mitochondrial steady-state transcripts in maize
- PMID: 30585757
- PMCID: PMC6380311
- DOI: 10.1080/15476286.2018.1561604
Major contribution of transcription initiation to 5'-end formation of mitochondrial steady-state transcripts in maize
Abstract
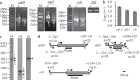
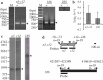
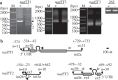
In plant mitochondria, some steady-state transcripts contain primary 5' ends derived from transcription initiation, while the others have processed 5' termini generated by post-transcriptional processing. Differentiation and mapping of the primary and processed transcripts are important for unraveling the molecular mechanism(s) underlying transcription and transcript end maturation. However, previous efforts to systematically differentiate these two types of transcripts in plant mitochondria failed. At present, it is considered that the majority of mature mRNAs may have processed 5' ends in Arabidopsis. Here, by combination of circular RT-PCR, quantitative RT-PCR, RNA 5'-polyphosphatase treatment and Northern blot, we successfully discriminated and mapped the primary and processed transcripts in maize mitochondria. Among the thirty-five mature and eight precursor RNAs analyzed in this study, about one half (21/43) were found to have multiple isoforms. In total, seventy-seven steady-state transcripts were determined, and forty-seven of them had primary 5' ends. Most transcription initiation sites (126/167) were downstream of a crTA-motif. These data suggested a major contribution of transcription initiation to 5'-end formation of steady-state transcripts in maize mitochondria. Moreover, the mapping results revealed that mature RNA termini had largely been formed before trans-splicing, and C→U RNA editing was accompanied with trans-splicing and transcript end formation in maize mitochondria.
Keywords: 5ʹ-end formation; RNA termini; maize; mature RNA; mitochondrion; precursor RNA; transcription initiation.
Figures


Similar articles
-
Discrimintion and Mapping of the Primary and Processed Transcripts in Maize Mitochondrion Using a Circular RT-PCR-based Strategy.J Vis Exp. 2019 Jul 29;(149). doi: 10.3791/60019. J Vis Exp. 2019. PMID: 31403631
-
RNA processing in plant mitochondria is independent of transcription.Plant Mol Biol. 2009 Aug;70(6):663-8. doi: 10.1007/s11103-009-9498-6. Epub 2009 May 3. Plant Mol Biol. 2009. PMID: 19412686
-
Transcription and RNA-processing in fission yeast mitochondria.RNA. 2005 May;11(5):785-95. doi: 10.1261/rna.7252205. Epub 2005 Apr 5. RNA. 2005. PMID: 15811919 Free PMC article.
-
Maturation of 5' ends of plant mitochondrial RNAs.Physiol Plant. 2016 Jul;157(3):280-8. doi: 10.1111/ppl.12423. Epub 2016 Mar 23. Physiol Plant. 2016. PMID: 26833432 Review.
-
From gene to protein in higher plant mitochondria.C R Acad Sci III. 2001 Mar;324(3):209-17. doi: 10.1016/s0764-4469(00)01293-2. C R Acad Sci III. 2001. PMID: 11291307 Review.
Cited by
-
Mitochondrial RNA maturation.RNA Biol. 2024 Jan;21(1):28-39. doi: 10.1080/15476286.2024.2414157. Epub 2024 Oct 10. RNA Biol. 2024. PMID: 39385590 Free PMC article. Review.
-
Mitochondrial mRNA Processing in the Chlorophyte Alga Pediastrum duplex and Streptophyte Alga Chara vulgaris Reveals an Evolutionary Branch in Mitochondrial mRNA Processing.Plants (Basel). 2021 Mar 18;10(3):576. doi: 10.3390/plants10030576. Plants (Basel). 2021. PMID: 33803683 Free PMC article.
-
Comparative analysis of mitochondrial genomes of maize CMS-S subtypes provides new insights into male sterility stability.BMC Plant Biol. 2022 Oct 1;22(1):469. doi: 10.1186/s12870-022-03849-6. BMC Plant Biol. 2022. PMID: 36180833 Free PMC article.
-
Mitochondrion-encoded circular RNAs are widespread and translatable in plants.Plant Physiol. 2022 Jun 27;189(3):1482-1500. doi: 10.1093/plphys/kiac143. Plant Physiol. 2022. PMID: 35325205 Free PMC article.
-
A linear and circular dual-conformation noncoding RNA involved in oxidative stress tolerance in Bacillus altitudinis.Nat Commun. 2023 Sep 15;14(1):5722. doi: 10.1038/s41467-023-41491-4. Nat Commun. 2023. PMID: 37714854 Free PMC article.
References
-
- Neupert W. Mitochondrial gene expression: a playground of evolutionary tinkering. Annu Rev Biochem. 2016;85:65–76. - PubMed
-
- Gualberto JM, Mileshina D, Wallet C, et al. The plant mitochondrial genome: dynamics and maintenance. Biochimie. 2014;100:107–120. - PubMed
-
- Choi BY, Acero MM, Bonen L. Mapping of wheat mitochondrial mRNA termini and comparison with breakpoints in DNA homology among plants. Plant Mol Biol. 2012;80:539–552. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
