Genetic diversity of Escherichia coli in gut microbiota of patients with Crohn's disease discovered using metagenomic and genomic analyses
- PMID: 30587114
- PMCID: PMC6307143
- DOI: 10.1186/s12864-018-5306-5
Genetic diversity of Escherichia coli in gut microbiota of patients with Crohn's disease discovered using metagenomic and genomic analyses
Abstract
Background: Crohn's disease is associated with gut dysbiosis. Independent studies have shown an increase in the abundance of certain bacterial species, particularly Escherichia coli with the adherent-invasive pathotype, in the gut. The role of these species in this disease needs to be elucidated.
Methods: We performed a metagenomic study investigating the gut microbiota of patients with Crohn's disease. A metagenomic reconstruction of the consensus genome content of the species was used to assess the genetic variability.
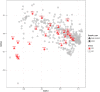
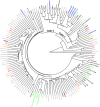
Results: The abnormal shifts in the microbial community structures in Crohn's disease were heterogeneous among the patients. The metagenomic data suggested the existence of multiple E. coli strains within individual patients. We discovered that the genetic diversity of the species was high and that only a few samples manifested similarity to the adherent-invasive varieties. The other species demonstrated genetic diversity comparable to that observed in the healthy subjects. Our results were supported by a comparison of the sequenced genomes of isolates from the same microbiota samples and a meta-analysis of published gut metagenomes.
Conclusions: The genomic diversity of Crohn's disease-associated E. coli within and among the patients paves the way towards an understanding of the microbial mechanisms underlying the onset and progression of the Crohn's disease and the development of new strategies for the prevention and treatment of this disease.
Keywords: Crohn’s disease; Escherichia coli; Gene content; Gut microbiota; Inflammatory bowel diseases; Metagenomics; Pangenome.
Conflict of interest statement
Ethics approval and consent to participate
This study was approved by the ethics committees of the Moscow Clinical Scientific Center and State Scientific Center of Coloproctology, Moscow, Russian Federation and carried out following the rules of the Declaration of Helsinki of 1975. All patients provided written informed consent for the sample collection and personal data processing.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Paramsothy S, Paramsothy R, Trainee G, Rubin DT, Kamm MA, Kaakoush NO, et al. Faecal Microbiota Transplantation for Inflammatory Bowel Disease: A Systematic Review and Meta-analysis. J Crohns Colitis. 11(10):1180-1199. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

