Comparative analysis of the mitochondrial genomes of oriental spittlebug trible Cosmoscartini: insights into the relationships among closely related taxa
- PMID: 30587118
- PMCID: PMC6307326
- DOI: 10.1186/s12864-018-5365-7
Comparative analysis of the mitochondrial genomes of oriental spittlebug trible Cosmoscartini: insights into the relationships among closely related taxa
Abstract
Background: Cosmoscartini (Hemiptera: Cercopoidea: Cercopidae) is a large and brightly colored Old World tropical tribe, currently containing over 310 phytophagous species (including some economically important pests of eucalyptus in China) in approximately 17 genera. However, very limited information of Cosmoscartini is available except for some scattered taxonomic studies. Even less is known about its phylogenetic relationship, especially among closely related genera or species. In this study, the detailed comparative genomic and phylogenetic analyses were performed on nine newly sequenced mitochondrial genomes (mitogenomes) of Cosmoscartini, with the purpose of exploring the taxonomic status of the previously defined genus Okiscarta and some closely related species within the genus Cosmoscarta.
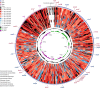
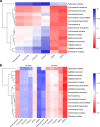
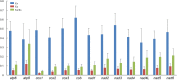
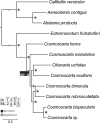
Results: Mitogenomes of Cosmoscartini display similar genomic characters in terms of gene arrangement, nucleotide composition, codon usage and overlapping regions. However, there are also many differences in intergenic spacers, mismatches of tRNAs, and the control region. Additionally, the secondary structures of rRNAs within Cercopidae are inferred for the first time. Based on comparative genomic (especially for the substitution pattern of tRNA secondary structure) and phylogenetic analyses, the representative species of Okiscarta uchidae possesses similar structures with other Cosmoscarta species and is placed consistently in Cosmoscarta. Although Cosmoscarta bimacula is difficult to be distinguished from Cosmoscarta bispecularis by traditional morphological methods, evidence from mitogenomes highly support the relationships of (C. bimacula + Cosmoscarta rubroscutellata) + (C. bispecularis + Cosmoscarta sp.).
Conclusions: This study presents mitogenomes of nine Cosmoscartini species and represents the first detailed comparative genomic and phylogenetic analyses within Cercopidae. It is indicated that knowledge of mitogenomes can be effectively used to resolve phylogenetic relationships at low taxonomic levels. Sequencing more mitogenomes at various taxonomic levels will also improve our understanding of mitogenomic evolution and phylogeny in Cercopidae.
Keywords: Cosmoscartini; Mitochondrial genome; Phylogeny; Spittlebug.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

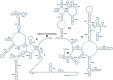
References
-
- Zhang DX, Hewitt GM. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 1997;25(2):99–120.
-
- Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410(2):103–123. - PubMed
-
- Curole JP, Kocher TD. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol. 1999;14(10):394–398. - PubMed
-
- Lin CP, Danforth BN. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol Phylogenet Evol. 2004;30(3):686–702. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

